Use {disk.frame} for your larger-than-RAM data manipulation needs:
* no longer be limited by the amt of RAM
* be able to use familiar tools such as dplyr verbs and data.table syntax
See diskframe.com
#rdiskframe #diskframe #rstats
Have a large CSV? `csv_to_disk.frame` is your friend:
* if you CSV is too large, consider setting in_chunk_size = 1e6 to load chunk by chunk
* a vector of CSV files as 1st argument to read them all
* uses data.table::fread, so is fast!
#rdiskframe
If you use RStudio, you can use <tab> to auto-complete disk.frame column names for you.
For more #diskframe convenience features, see diskframe.com/articles/conve…
#rstats

A disk.frame is made up of chunks. Each chunk is a .fst file. The most common (and default) naming pattern is 1.fst, 2.fst etc. This is not strict.
`get_chunk(df, n)` to get nth chunk and test the algo before applying to full disk.frame
#rstats
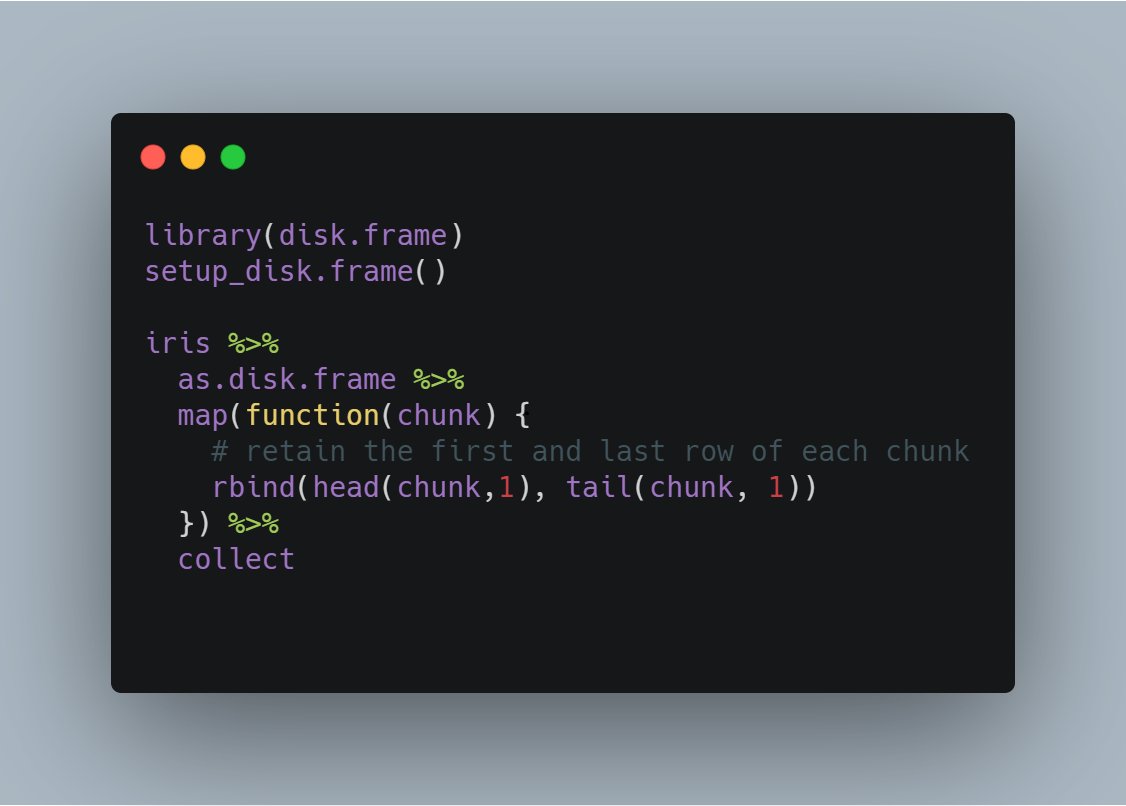
`map`, from {purrr}, is how I envisioned {disk.frame} at its best
In #diskframe context, `map(df, fn)` applies the `fn` to each chunk of `df`
Best way to apply arbitrary R-native user-defined functions to each chunk of a disk.frame
#rstats
Use `delete(df)` to delete a disk.frame
Use `move_to(df, new_path)` to move a disk.frame
#rstats #diskframe
`map` applies the same function to each chunk. You want a more convenient way?
Use `create_chunk_mapper` e.g.
filter <- create_chunk_mapper(dplyr::filter)
That's how {disk.frame}'s filter is defined!
#rstats #diskframe
Let `cpun` = number of CPU cores
{disk.frame} is very efficient if the
`cpun` * (RAM required for one chunk) = 70%~80% RAM
because #diskframee loads `cpun` chunks into RAM in parallel, and loading as much data in one go = faster
#rstats
When using `csv_to_disk.frame` the # of chunks is calculated using a simple formula in `recommend_nchunks`
The formula is based on
* RAM size - more RAM less chunks
* # of CPU cores - more cores more chunks, and
* the size of your CSV
#rstats
If you move a disk.frame to a new computer with more RAM and more CPU cores? You can use the `rechunk` function to inc or dec the number of chunks.
RAM doubled? Then 1/2 the # of chunks
CPU cores doubled? Then double the # of chunks too
#rstats
`delayed(df, fn)` applies an arbitrary function to `df` lazily
The mechanism via which {disk.frame} does lazy is by storing the `fn` in the attribute attr(df, "lazy_fn"). It also captures any relevant environment variables used in `fn`
#rstats
`df %>% dplyr::some_func %>% delalyed(fn)` records two functions in the `attr(df, "lazy_fn")` and the `disk.frame:::play` runs them when `collect` is called
So the mechanism is to record a list of functions to be played. Inspired by {chunked}
Consider
`df %>%
fn1 %>%
fn2 %>%
collect`
`fn1` and `fn2` will be executed on each chunk of `df` via the play mechanism. The results are transferred back to the main session.
Try to minimize the amt of transfers, because it can be slow
{disk.frame} follows a very simplified form of map-reduce, in that the map phase is applied to chunk and the reduce phase is simply to row-bind of results.
Actually, the map phase can produce non-data.frames, but then there are two limitations TBC
In the map phase, we can produce non-data.frames, but two limitations
1. it cannot be saved to disk with `write_disk.frame`. In fact, any format not compatible with fst format is not allowed
2. `collect` will return a list
#rstats
{disk.frame} has implemented left/inner/full/anti/semi_join.
The only join missing is right_join; just use left_join.
xxx_join(df, df2) works best when the df2 is a data.frame instead of a disk.frame
#rstats
xxx_join(df, df2) if both df and df2 are disk.frame then to implement the join algorithm properly would be very expensive and slow!
For now, you can only merge by matching chunk IDs, i.e. only those with same chunk id will be merged
#rstats
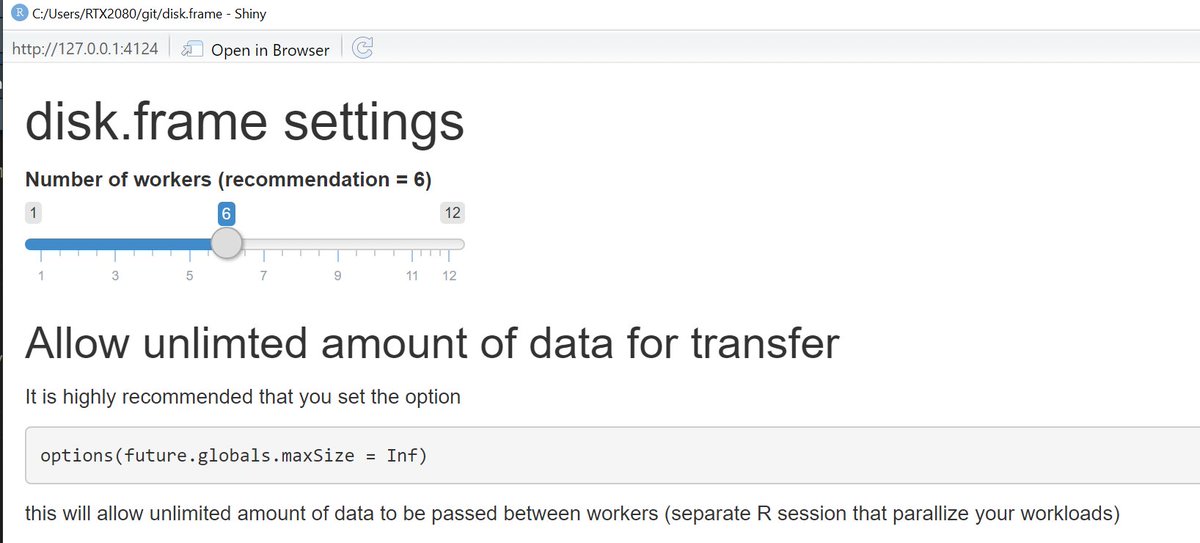
No one can remember all the options a package offers. The list of options for {disk.frame} is small right now, but a GUI for options is still good UX, IMHO.
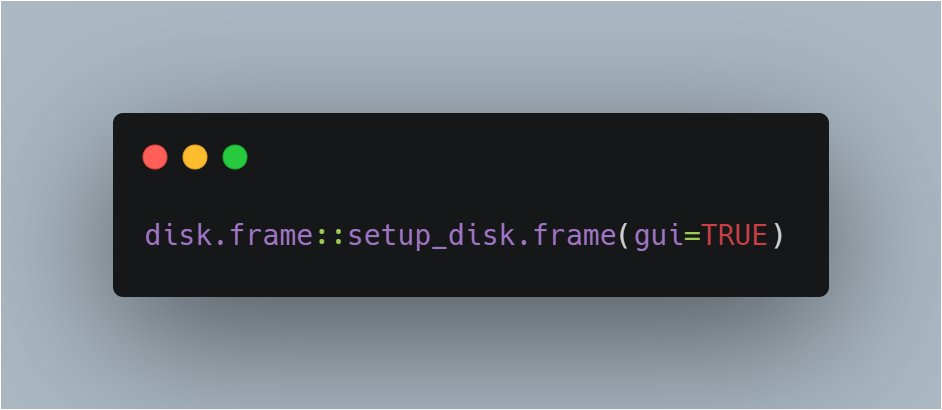
`disk.frame::setup_disk.frame(gui=TRUE)`
will open a Shiny app for options.
#rstats
1-stage group-by is possible from v0.3
df %>% group_by(g) %>% summarize(mean(x))
Define custom group_by summarization
`fn_df.chunk_agg.disk.frame` = chunk-lvl
`fn_df.collected_agg.disk.frame` = post-collect
See diskframe.com/articles/11-cu… #rstats
Only 1 take-away? Make it
`srckeep`
"keep" only these columns at "source"
df %>%
srckeep(c("g", "x")) %>%
group_by(g) %>%
summarize(mean(x))
will use {fst}`s random column access to load just cols g and x into RAM = much FASTER
#rstats
{disk.frame} is purrr friendly. You can apply a function to each chunk using the cmap family of functions like cimap, cmap_dfr etc
The c is for chunk, cmap_dfr row binds the results just like map_dfr.
E.g.
cimap(df, ~fn(.x,.y))
#rstats
1.
Forgot {disk.frame} boilerplate code? Use `show_boilerplate()`
2.
{disk.frame} follows semver so v0.2.0 --> v0.3.0 means breaking API. Expect 1 round of deprecation warnings
data.table syntax API is likely to change soon
#rstats
`shard(df, shard_by = "col1"))` will distribute the rows of `df` so that rows with same value in column `col1` will end up in the same chunk.
This is great for group_by `col1`
df %>%
chunk_group_by(col1) %>%
chunk_summarize(...)
#rstats
Most functions that write a disk.frame to disk defaults to writing to the `tempdir()`.
For datasets, the user wants to re-use, they should use the `outdir = your_path` argument to store it somewhere permanent.
#rstats
{disk.frame} wouldn't have been possible without the awesomeness of
fst fstpackage.org
future github.com/HenrikBengtsso…
dplyr dplyr.tidyverse.org/articles/progr…
data.table #rdatatable
Rcpp github.com/RcppCore/Rcpp
Thank you all!
#rstats
github.com/sponsors/eddel…
This is the link to the list of developers sponsored by @rstudio on Github github.com/rstudio-sponso…
fst and future do not have sponsorship links.


