Since some RNA mutagens "could induce the same mutation pattern", authors suggest further investigation.
biorxiv.org/content/10.110…
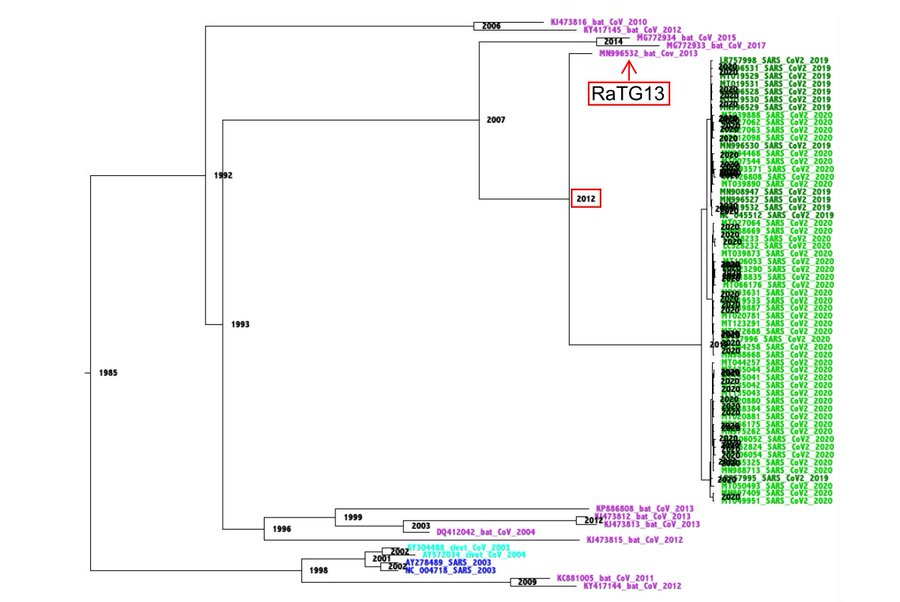
Issue: two bat CoVs show nonsyn mutations evenly distributed along their S sequences. For RaTG13 vs SARS-CoV-2 they appear heavily concentrated around codons 300-500. Why?
nerdhaspower.weebly.com/ratg13-is-fake…
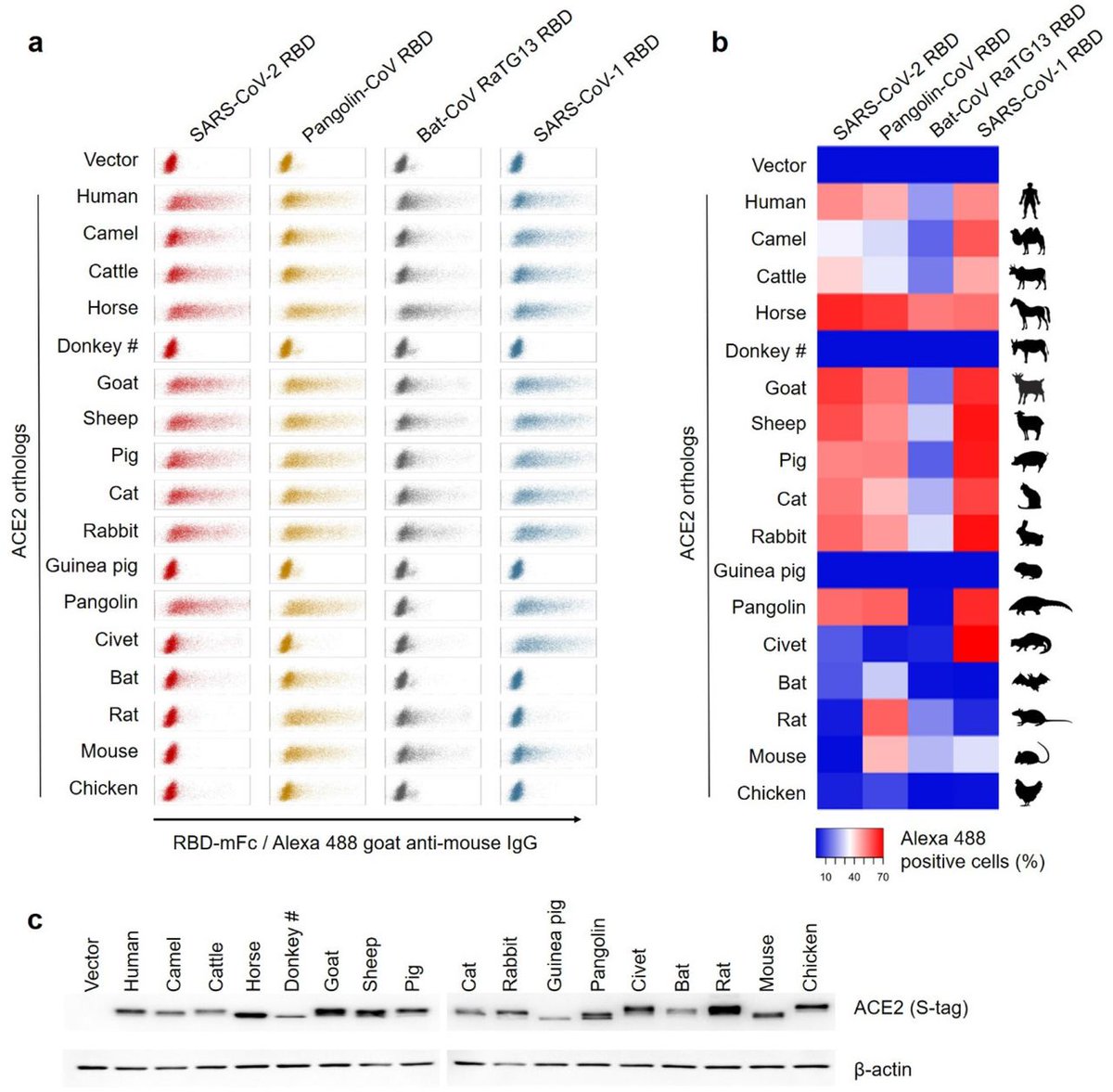
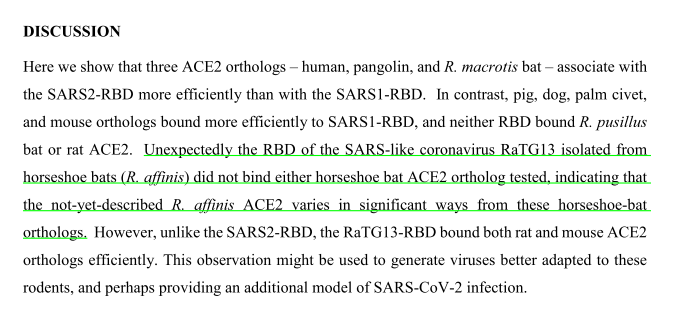
A study found this synthetic spike showing a surprisingly low affinity for usually tested ACE2, and for bat ACE2 in particular. biorxiv.org/content/10.110…
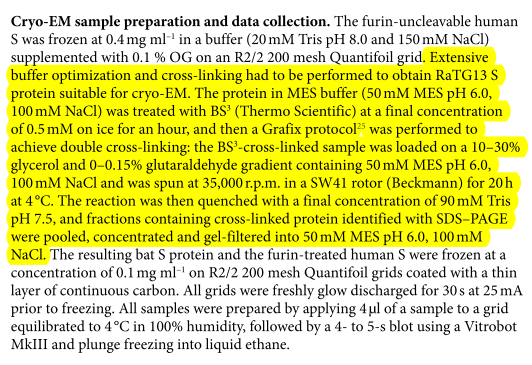
They report that RaTG13 spike protein was found to be so unstable that it required extensive "optimization and cross-linking" in order to be micrographed. nature.com/articles/s4159…
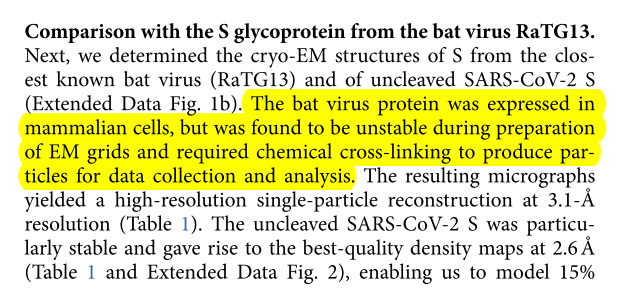
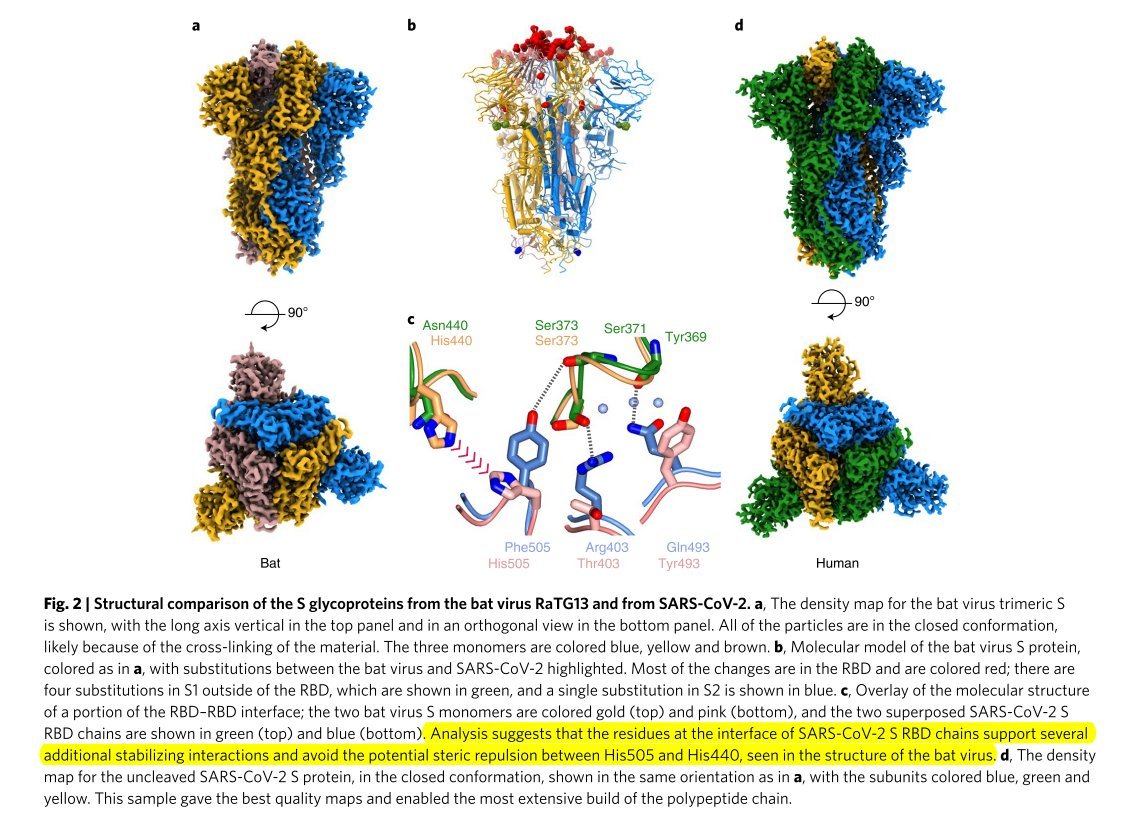
This comparison was much lower for SARS-CoV and non-existent for HKU1.
peerj.com/articles/9648/
Based on a protein metric, MeaPED, @some_wise estimates they split in 2012.
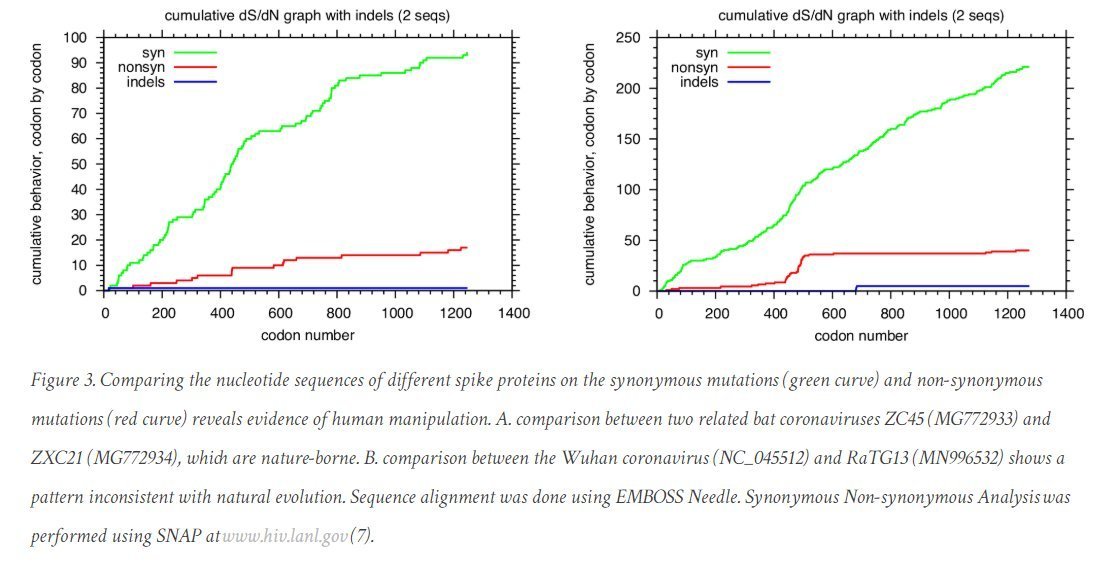
Another sign of dN/dS artifacts in RaTG13 sequence?
osf.io/ntc8w