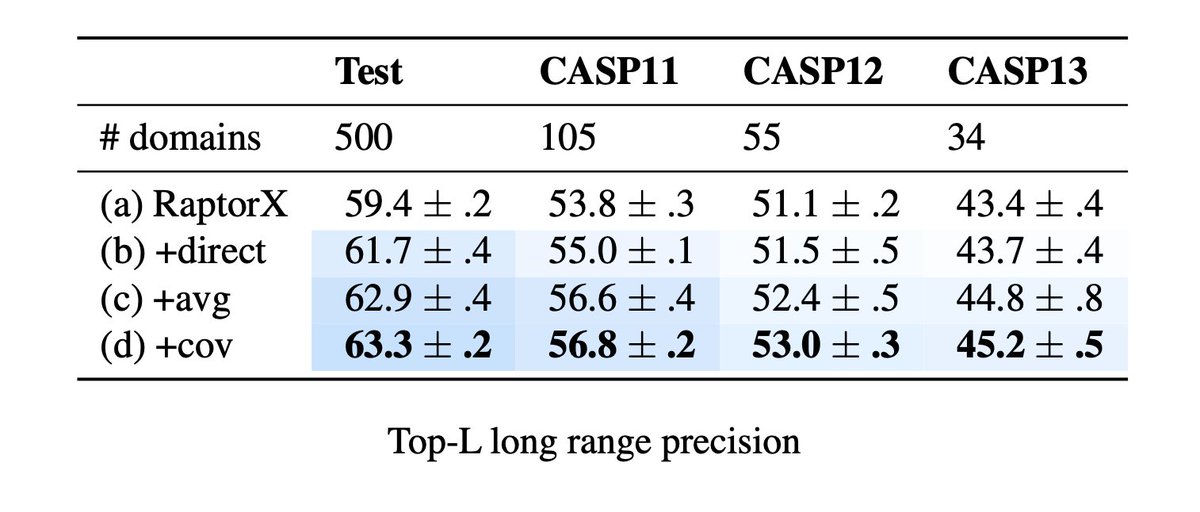
1/9 Today we’re excited to release Transformer models pre-trained on evolutionary-scale protein sequence data along with a major update to our preprint from last year:
Paper: biorxiv.org/content/10.110…
Models: github.com/facebookresear…
Paper: biorxiv.org/content/10.110…
Models: github.com/facebookresear…
2/9 We added extensive new benchmarks for remote homology, secondary structure, long range contacts, and mutational effect. Improvements to downstream models lead to SOTA features across multiple benchmarks.
3/9 There are two larger questions we’re interested in answering: (1) can language models learn biology from sequences; (2) are there favorable scaling laws for data and model parameters, i.e. similar to those observed in NLP. In new work we find support for both.
4/9 Last year in the first version of the paper, we scaled Transformer models with ~700M parameters to 250M protein sequences in UniParc. The models learn about the intrinsic properties of proteins. 
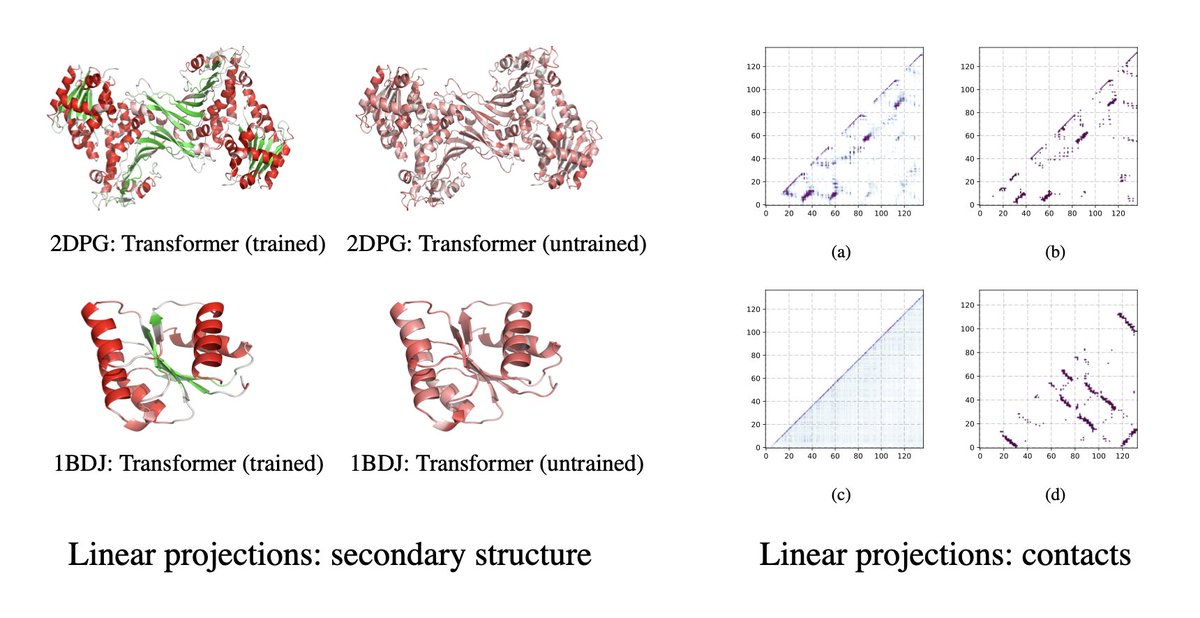
5/9 In new experiments we look at differences between datasets (UniRef50 vs UniRef100), model architectures (LSTMs vs Transformers), and parameters (small vs large Transformers).
Transformer architectures (vs LSTM), diversity in data, and scale in parameters have big impact.
Transformer architectures (vs LSTM), diversity in data, and scale in parameters have big impact.
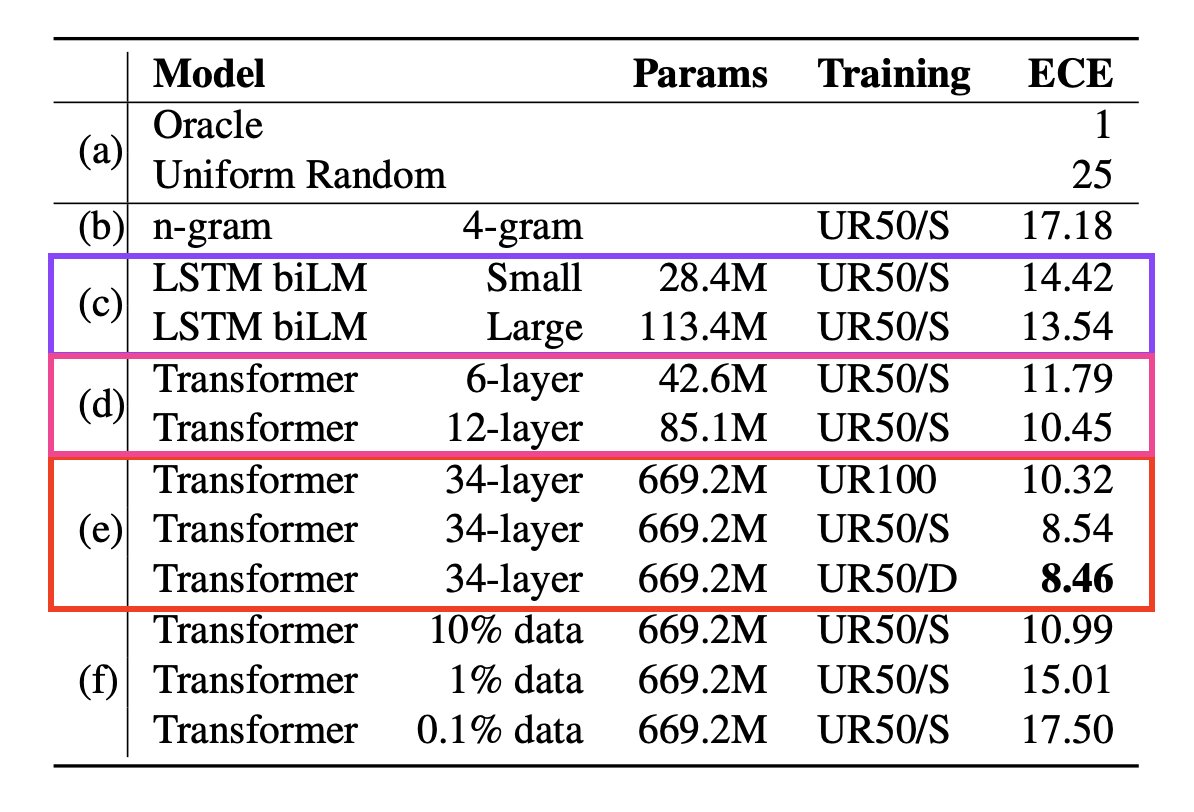
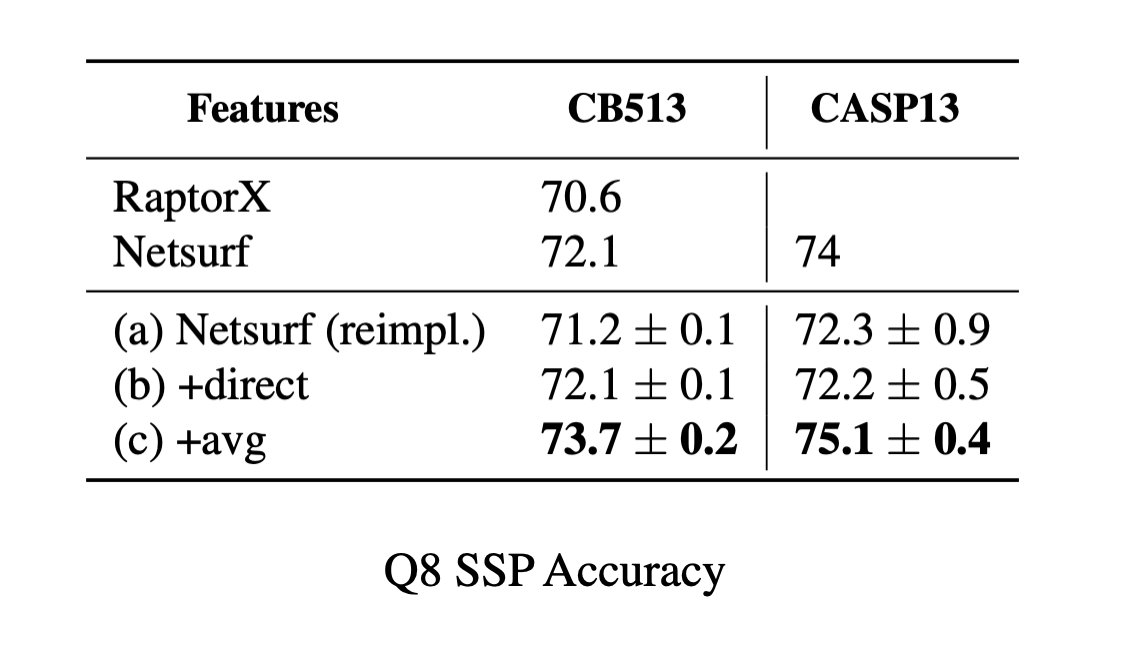
6/9 Combining features from representation learning with features used in SOTA structure prediction methods improves performance. Example, secondary structure prediction: 
8/9 Other great work investigating representation learning for protein sequences:
UniRep. biorxiv.org/content/10.110…
SeqVec. biorxiv.org/content/10.110…
TAPE. arxiv.org/abs/1906.08230
UniRep. biorxiv.org/content/10.110…
SeqVec. biorxiv.org/content/10.110…
TAPE. arxiv.org/abs/1906.08230
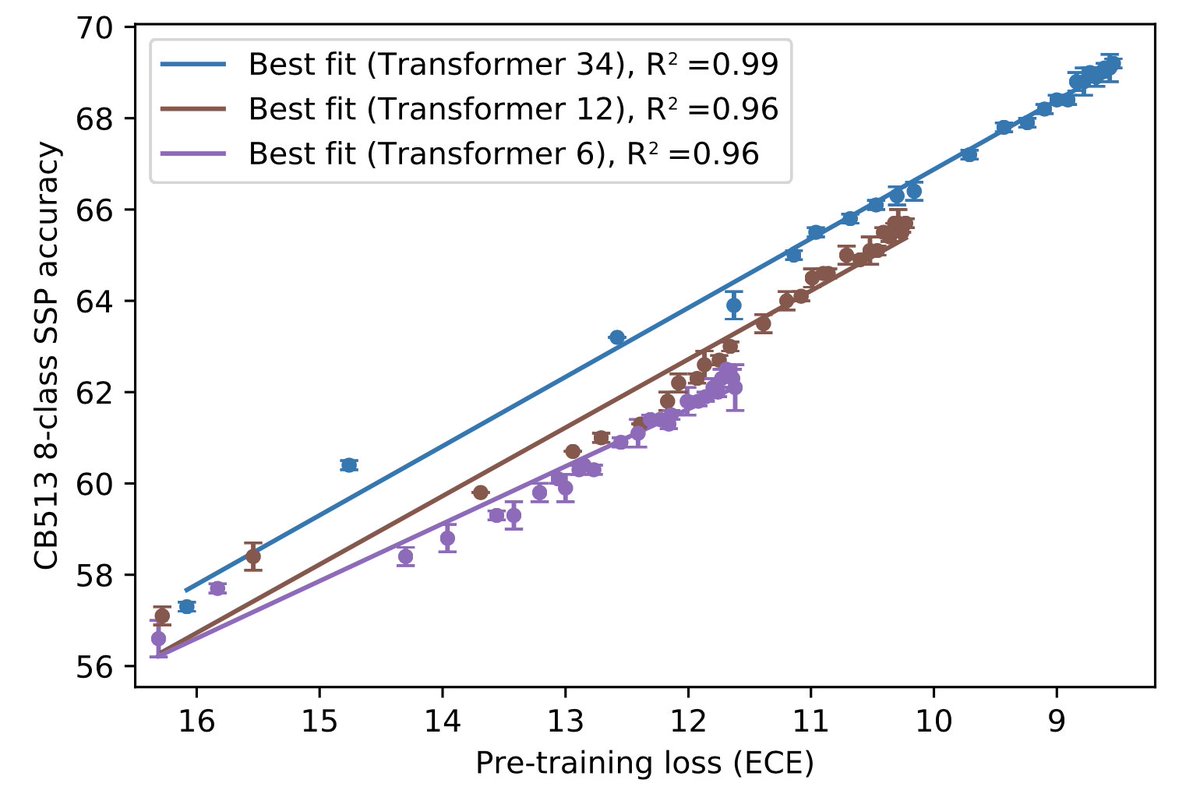
9/9 A first answer to the question about scaling laws. Relationship between language modeling fidelity and downstream performance is linear over the course of training! Suggests results will continue to improve with scale. 
• • •
Missing some Tweet in this thread? You can try to
force a refresh