Does gene expression predict wiring in the cortex?
Here we introduce a new technique, BARseq2, to answer this question
BARseq2 looks at genes and axon projections at high throughput--thousands of single neurons and many genes per cell in parallel
doi.org/10.1101/2020.0…
1/n
Here we introduce a new technique, BARseq2, to answer this question
BARseq2 looks at genes and axon projections at high throughput--thousands of single neurons and many genes per cell in parallel
doi.org/10.1101/2020.0…
1/n
This is work by an amazing team: Yu-Chi Sun, Xiaoyin Chen, Stephan Fischer, @starfishlu , and Jesse Gillis
(2/n)
(2/n)
BARseq2 builds on first-generation BARseq (cell.com/cell/pdf/S0092…), which let us look at barcoded axonal projections by in situ DNA sequencing
BARseq2 adds highly multiplexed sequencing of dozens of endogenous genes
shoulders of giants: @MatsNilssonLab @jlee8usa
(3/n)
BARseq2 adds highly multiplexed sequencing of dozens of endogenous genes
shoulders of giants: @MatsNilssonLab @jlee8usa
(3/n)

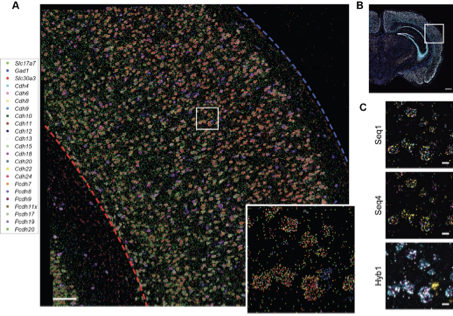
We hypothesized that cadherin expression was key. So we used BARseq2 to detect cadherin expression and cell type markers in auditory and motor cortex
(4/n)
(4/n)
Laminar patterns of gene expression obtained by BARseq2 were similar to the that seen in the Allen Brain Atlas
(5/n)
(5/n)
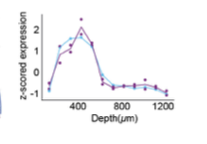
The sensitivity of BARseq2 was similar to 10x v3 in this particular experiment, but if we push it we can reach at least 60% of RNAscope
(6/n)
(6/n)

We also determined transcriptomic cell types by detecting additional cell type markers in the motor cortex.
7/n
7/n

Combining in situ sequencing of barcodes and endogenous genes, we correlated cadherin expression to projections in 1,349 single neurons in auditory and motor cortex.
Gene expression+single neuron axon projection patterns in 1000 neurons, from just a handful of animals!
8/n
Gene expression+single neuron axon projection patterns in 1000 neurons, from just a handful of animals!
8/n

Our data recapitulated known differences in gene expression and projections between neuronal classes, types, and subtypes.
For example, neurons that project only ipsilaterally were mostly IT4 transcriptomic type (i.e. L6 Car3 type)
9/n
For example, neurons that project only ipsilaterally were mostly IT4 transcriptomic type (i.e. L6 Car3 type)
9/n

How do these cadherins relate to transcriptomic cell types?
To find out, we grouped cadherins into co-expression modules and found that these modules were associated with multiple branches of the transcriptomic taxonomy
10/n
To find out, we grouped cadherins into co-expression modules and found that these modules were associated with multiple branches of the transcriptomic taxonomy
10/n
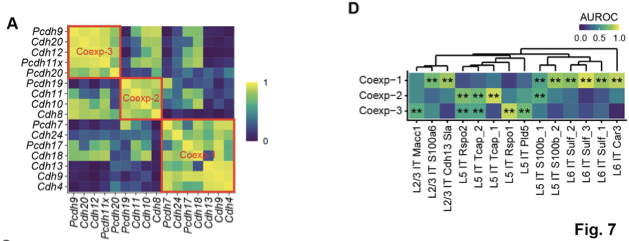
Our results suggest that the relationship between gene expression and axonal projections in the cortex is complex
Probably not fully described by the transcriptomic types alone
Maybe the key is to look during development? Or at other genes? Stay tuned...
11/11
Probably not fully described by the transcriptomic types alone
Maybe the key is to look during development? Or at other genes? Stay tuned...
11/11
• • •
Missing some Tweet in this thread? You can try to
force a refresh