Today Google @DeepMind announced that their deep learning system AlphaFold has achieved unprecedented levels of accuracy on the “protein folding problem”, a grand challenge problem in computational biochemistry.
What is this problem, and why is it hard?
deepmind.com/blog/article/a…
What is this problem, and why is it hard?
deepmind.com/blog/article/a…
I spent a couple years on this problem in a junior role in the early days of @DEShawResearch, so it's close to my heart. DESRES (as we called it internally) took an approach of building a specialized supercomputing architecture, called Anton: 

Proteins are long chains of amino acids. Your DNA encodes these sequences, and RNA helps manufacture proteins according to this genetic blueprint.
Proteins are synthesized as linear chains, but they don't stay that way. They fold up in to complex, globular shapes.
Proteins are synthesized as linear chains, but they don't stay that way. They fold up in to complex, globular shapes.

One part of the chain might coil up into a tight spiral called an α-helix. Another part might fold back and forth on itself to create a wide, flat piece called a β-sheet. 
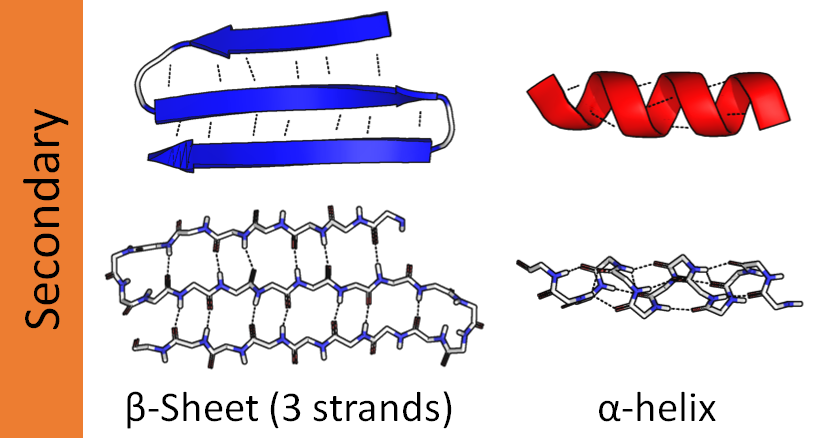
The sequence of amino acids itself is called primary structure. Components like this are called secondary structure.
Then, these components themselves fold up among themselves to create unique, complex shapes. This is called tertiary structure.
Then, these components themselves fold up among themselves to create unique, complex shapes. This is called tertiary structure.
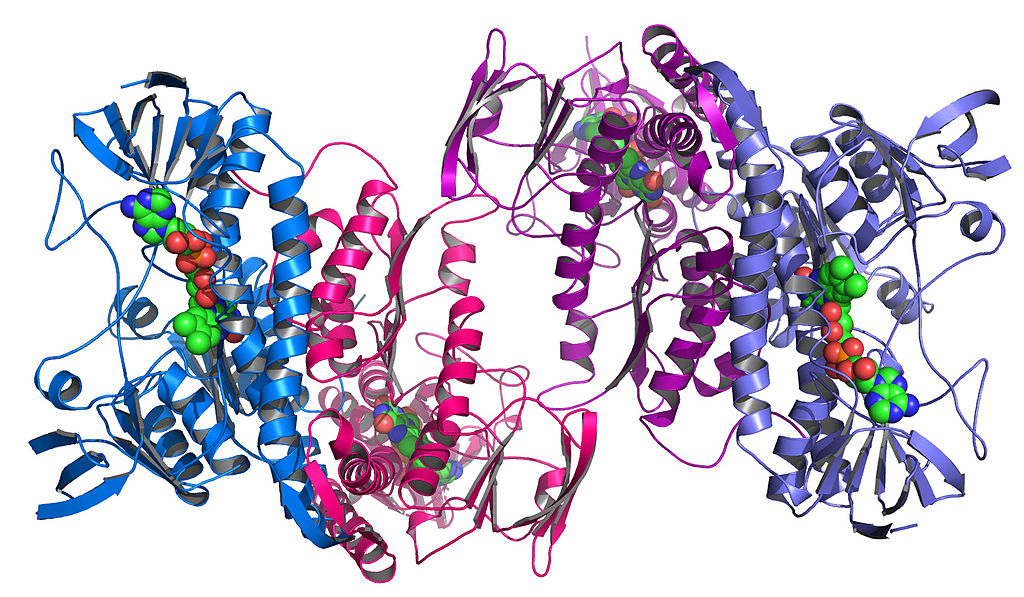
This looks like a mess. Why does this big tangle of amino acids matter?
Protein structure is not random! Each protein folds in a specific, unique, and largely predictable way that is essential to its function.
Protein structure is not random! Each protein folds in a specific, unique, and largely predictable way that is essential to its function.
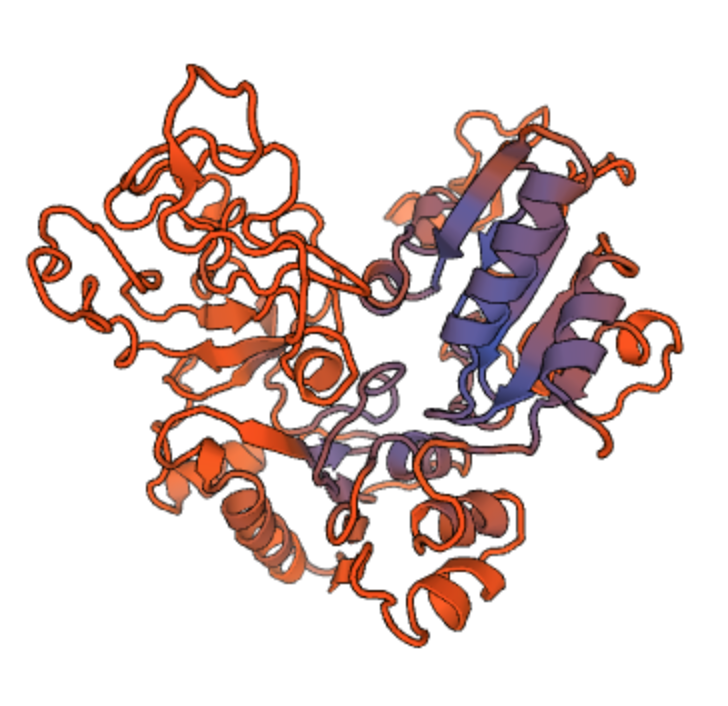
The physical shape of a protein gives it a good fit to targets it might bind with. Other physical properties matter too, especially the distribution of electrical charge within the protein, as shown here (positive charge in blue, negative in red): 

If a protein is essentially a self-assembling nanomachine, then the main purpose of the amino acid sequence is to produce the unique shape, charge distribution, etc. that determines the protein's function.
*How* exactly this happens, in the body, is still not fully understood, and is an active area of research. In any case, understanding structure is crucial to understanding function.
But the DNA sequence only gives us the primary structure of a protein. How can we learn its secondary and tertiary structure—the exact shape of the blob?
This problem is called “protein structure determination”, and there are two basic approaches: measurement and prediction.
This problem is called “protein structure determination”, and there are two basic approaches: measurement and prediction.
Experimental methods can measure protein structure. But it isn't easy: an optical microscope can't resolve the structures.
For a long time, X-ray crystallography was the main method. NMR has also been used, and more recently, a technique called cryogenic electron microscopy.
For a long time, X-ray crystallography was the main method. NMR has also been used, and more recently, a technique called cryogenic electron microscopy.
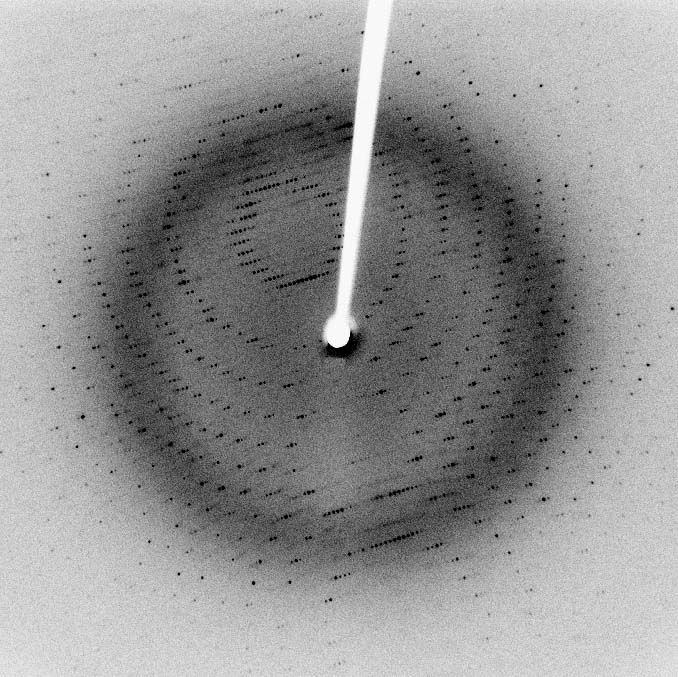
But these methods are difficult, expensive, and time-consuming, and they don't work for all proteins.
Notably, proteins embedded in the cell membrane—such as the ACE2 receptor that COVID-19 binds to—fold in the lipid bilayer of the cell and are difficult to crystallize.
Notably, proteins embedded in the cell membrane—such as the ACE2 receptor that COVID-19 binds to—fold in the lipid bilayer of the cell and are difficult to crystallize.
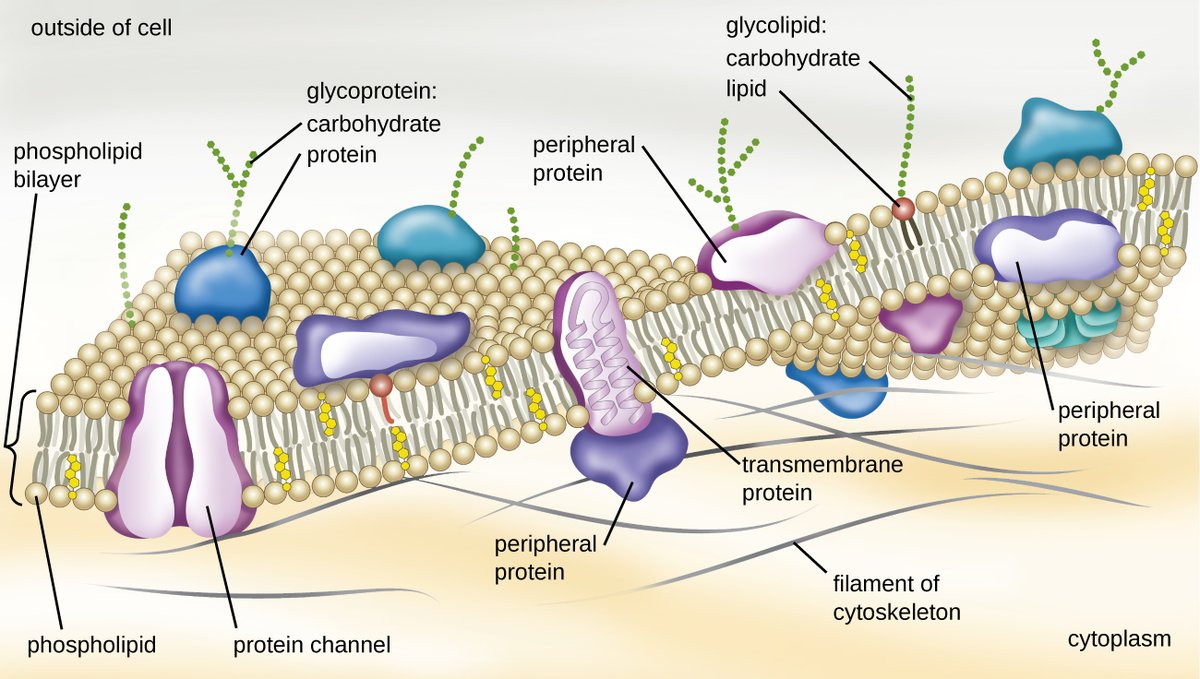
Because of this, we have only determined the structure of a tiny percentage of the proteins that we've sequenced. Google notes that there are 180M protein sequences in the Universal Protein database, but only ~170k structures in the Protein Data Bank.
We need a better method.
We need a better method.
Remember, though, that the secondary and tertiary structures are mostly a function of the primary structure, which we know from genetic sequencing.
What if, instead of *measuring* a protein's structure, we could *predict* it?
What if, instead of *measuring* a protein's structure, we could *predict* it?
This is “protein structure prediction”, or colloquially, the “protein folding problem.” Computational biochemists have been working on it for decades.
How could we approach this?
How could we approach this?
The obvious way is to directly simulate the physics. Model the forces on each atom, given its location, charge, and chemical bonds. Calculate accelerations and velocities based on that, and evolve the system step by step. This is called “molecular dynamics” (MD).
The problem is that this is *extremely* computationally intensive.
A typical protein has hundreds of amino acids, which means thousands of atoms. But the environment also matters: the protein interacts with surrounding water when folding. So it's more like 30k atoms to simulate.
A typical protein has hundreds of amino acids, which means thousands of atoms. But the environment also matters: the protein interacts with surrounding water when folding. So it's more like 30k atoms to simulate.
And there are electrostatic interactions between every pair of atoms, so naively that's ~450M pairs, an O(N²) problem. (There are smart algorithms to make this O(N log N).)
Also IIRC you end up needing to run for something like 10^9 to 10^12 timesteps.
It's a pain.
Also IIRC you end up needing to run for something like 10^9 to 10^12 timesteps.
It's a pain.
OK, but we don't have to simulate the entire folding process.
Another approach is to find the structure that *minimizes potential energy*. Objects tend to come to rest at energy minima, so this is a good heuristic. The same model that gives us forces for MD can calculate energy.
Another approach is to find the structure that *minimizes potential energy*. Objects tend to come to rest at energy minima, so this is a good heuristic. The same model that gives us forces for MD can calculate energy.
With this approach, we can try a whole bunch of candidate structures and pick the one with lowest energy.
The problem, of course, is where do you get the structures from? There are just way too many—molecular biologist Cyrus Levinthal estimated 10^300 (!)
The problem, of course, is where do you get the structures from? There are just way too many—molecular biologist Cyrus Levinthal estimated 10^300 (!)
Of course, you can be much smarter than trying all of them at random. But there are still too many.
So there have been many attempts to get faster at doing these kinds of calculations.
So there have been many attempts to get faster at doing these kinds of calculations.
Anton, the supercomputer from @DEShawResearch, used specialized hardware—a custom integrated circuit. IBM also has a computational bio supercomputer, Blue Gene. 

Stanford created Folding@Home to leverage the massively distributed power of ordinary home computers.
The Foldit project from UW makes folding a game, to augment computation with human intuition.
The Foldit project from UW makes folding a game, to augment computation with human intuition.
Still, for a long time, no technique was able to predict a wide variety of protein structures with high accuracy. A biannual competition called CASP, which compares algorithms against experimentally measured structures, saw top scores of 30–40%… until recently: 

So how does AlphaFold work? It uses multiple deep neural nets to learn different functions relevant to each protein. One key function is a prediction of the final *distances* between pairs of amino acids. This guides the algorithm to the final structure.
In one version of the algorithm, they then derived a potential function from this prediction, and applied simple gradient descent—which worked remarkably well. (I can't tell from what I've been able to read today if this is still what they're doing.)
A general advantage of AlphaFold over some previous methods is that it doesn't need to make assumptions about the structure. Some methods work by splitting the protein into regions, figuring out each region, then putting them back together. AlphaFold doesn't need to do this.
@DeepMind seems to be calling the protein folding problem solved, which strikes me as simplistic, but in any case this appears to be a major advance. Experts outside Google are calling it “fantastic”, “gamechanging”, etc.
sciencemag.org/news/2020/11/g…
sciencemag.org/news/2020/11/g…
Between protein folding and CRISPR, genetic engineering now has two very powerful new tools in its toolbox. Maybe the 2020s will be to biotech what the 1970s were to computing.
Congrats to the researchers at @DeepMind on this breakthrough!
Congrats to the researchers at @DeepMind on this breakthrough!
Blog post version of this thread, with image credits, at @rootsofprogress: rootsofprogress.org/alphafold-prot…
PS: the impact AlphaFold can have on pharmaceuticals
https://twitter.com/russelljkaplan/status/1485191271840837633
• • •
Missing some Tweet in this thread? You can try to
force a refresh













