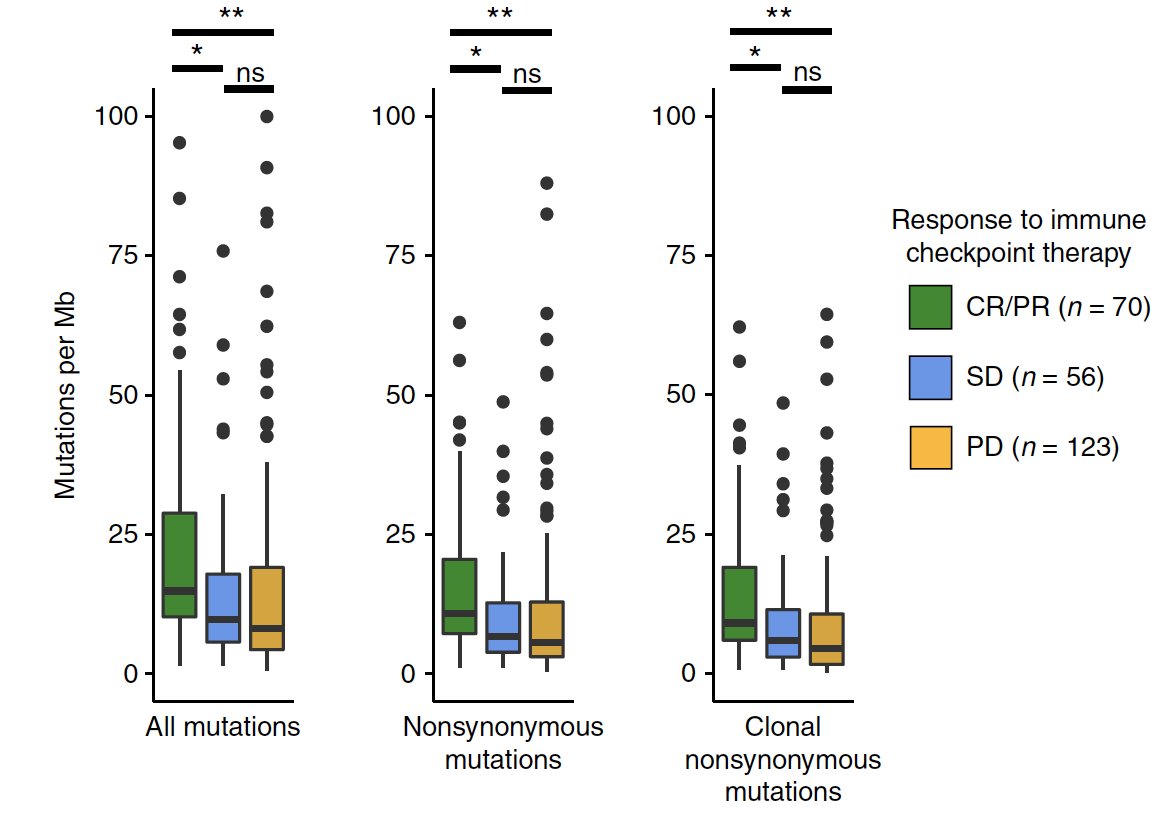
Here's our latest, a deep molecular dive into exceptional responders to anti-androgen therapy before surgery - congrats @aloktewar @alexorscanner Dr. Taplin @DanaFarber_GU et al!
cell.com/cell-reports/f… @CellReports
[1/n]
cell.com/cell-reports/f… @CellReports
[1/n]
(See also co-published/co-submitted wonderful story by @Chris_Barbieri1 lab with convergent science! cell.com/cell-reports/f…)
High risk localized prostate cancer can be curable with existing modalities (e.g. surgery), though whether giving upfront anti-androgen therapy before surgery can expand the number of men who are cured is not yet known.
Many trials ongoing by Dr. Taplin + many others to assess
Many trials ongoing by Dr. Taplin + many others to assess
This clinical context also allows for a unique opportunity to directly study the effect of these therapies in human tumors, where we can ask why some patients' tumors exhibit deep biological responses vs. persistent high risk disease at surgery 

Since the definition of "exceptional responder" will (of course) vary by study given the wide range of clinical contexts, therapies, and outcome measures in such work, for clarity in this study we defined these responses as follows: 
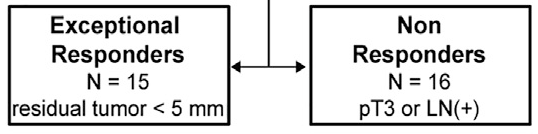
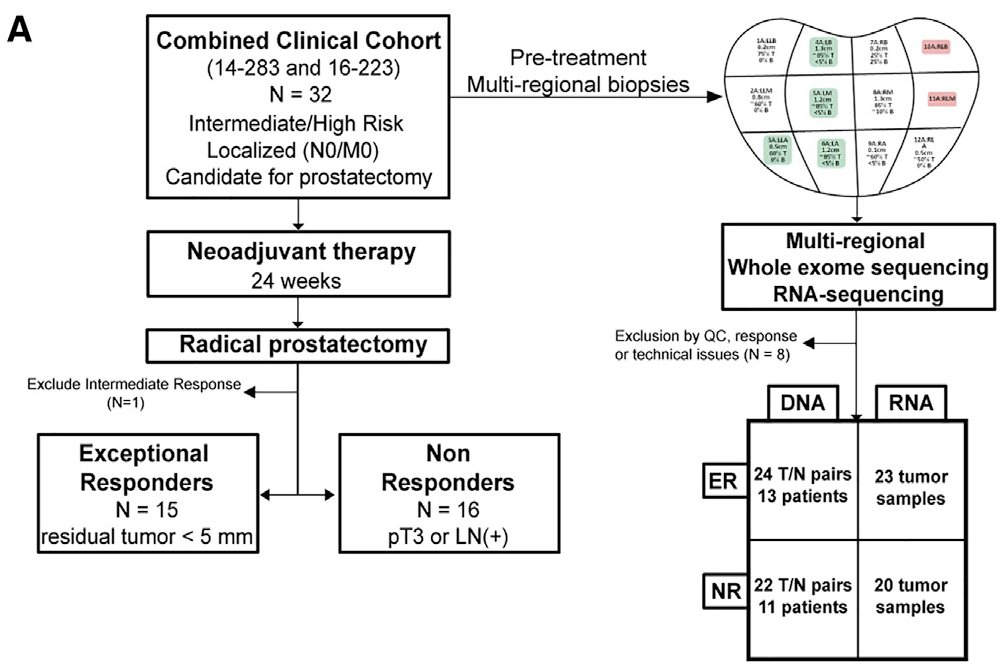
We accessed pre-treatment biopsy specimens (often more than one sample per-patient to evaluate genomic heterogeneity) from men enrolled on these clinical studies...
...and did a deep dive into molecular differences between response groups to identify potential explanations
...and did a deep dive into molecular differences between response groups to identify potential explanations
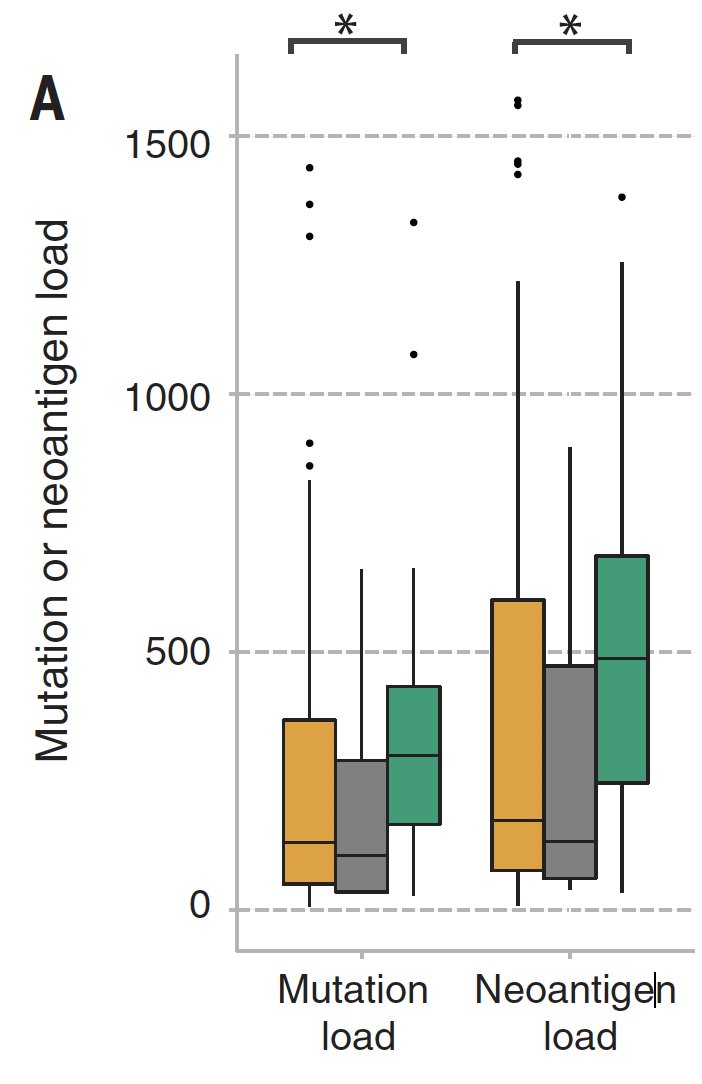
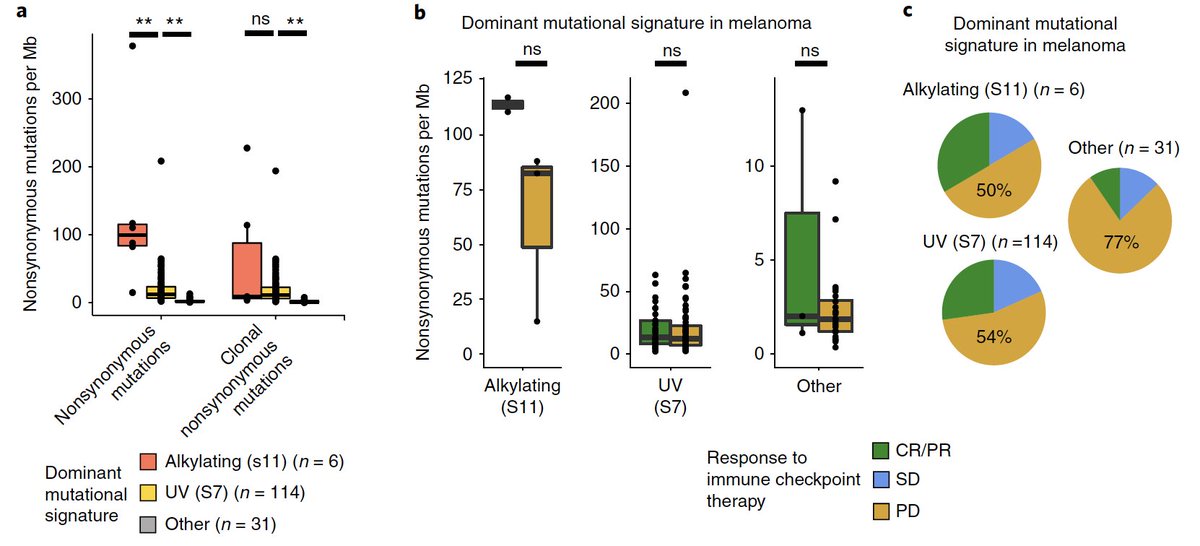
We found SPOP mutations (and, curiously, deletions in SPOPL - a paralog) highly enriched in patients with exceptional responses...
...whereas TP53 and PTEN mutations were enriched in nonresponders

...whereas TP53 and PTEN mutations were enriched in nonresponders
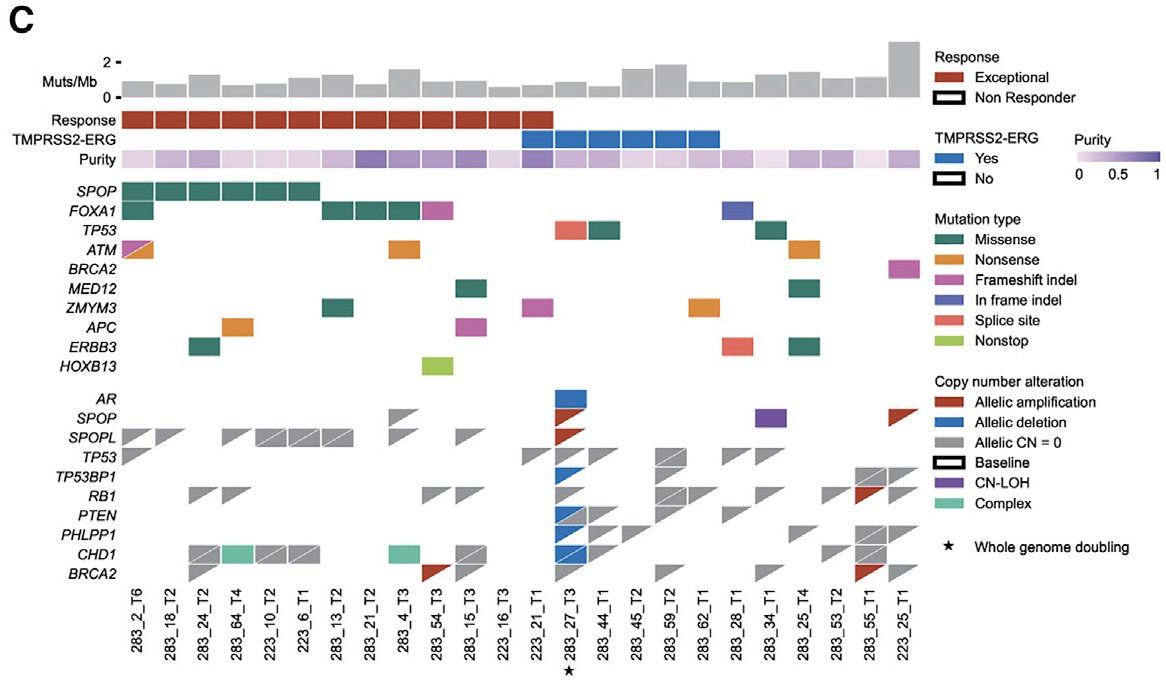
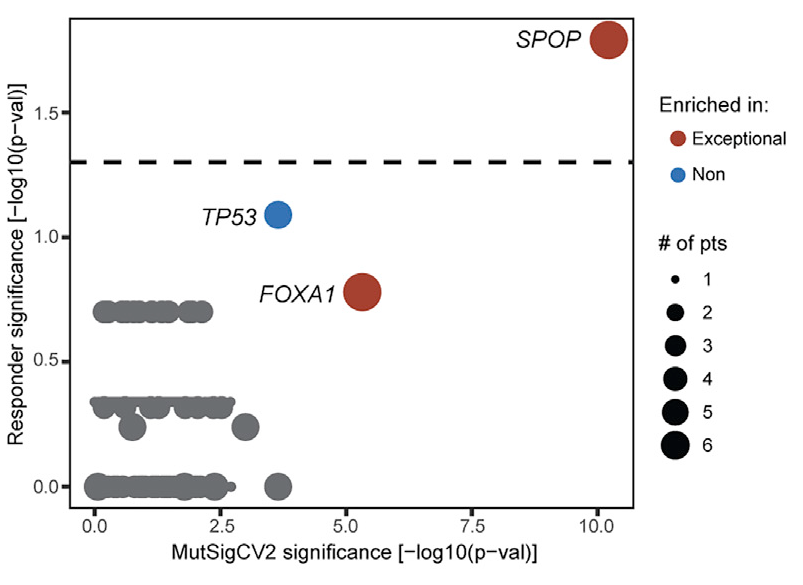
This ties nicely into the biological investigations into SPOP and androgen biology that @Chris_Barbieri1 and I discussed at @PCFnews Annual Meeting 2019 that led to cosubmission! It also relates to an amazing @sowalsky study where this trend was also seen europeanurology.com/article/S0302-…
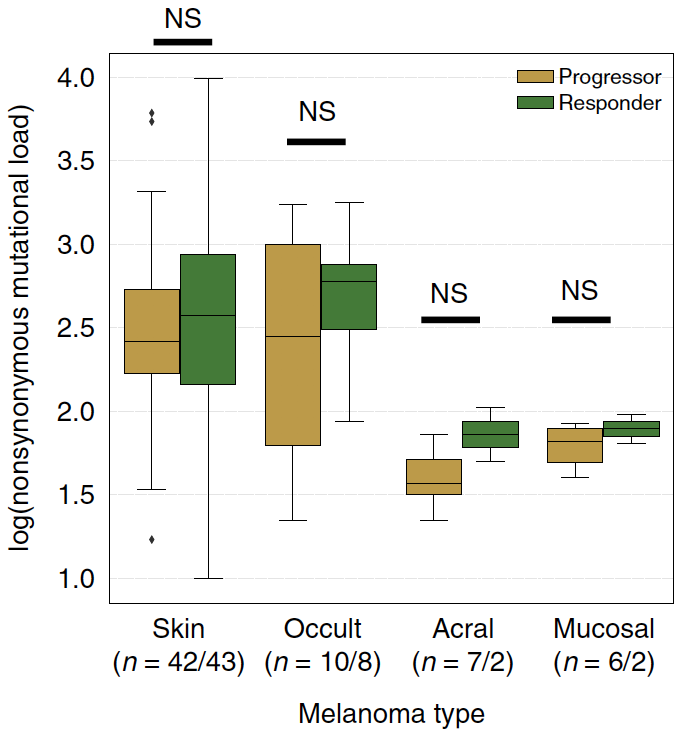
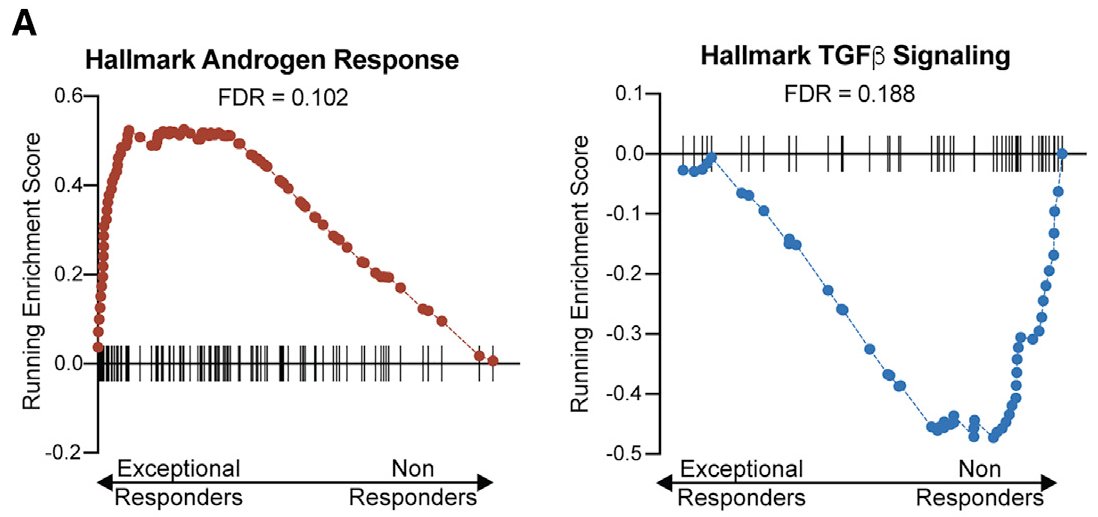
We also looked at transcriptional programs btwn clinical groups & noted androgen response elevated in responders (no surprise)...
...and TGF-beta signaling in nonresponders (see also: similar findings in metastatic prostate cancer h/t @MXresearch nature.com/articles/s4159…)
...and TGF-beta signaling in nonresponders (see also: similar findings in metastatic prostate cancer h/t @MXresearch nature.com/articles/s4159…)
Using multi-regional analysis, we also looked for tumor heterogeneity patterns in responders (h/t @getz_lab @gaddyg + co.)
We noted that interesting mutations in this treatment context were truncal, but couldn't find global patterns related to response in this cohort
We noted that interesting mutations in this treatment context were truncal, but couldn't find global patterns related to response in this cohort
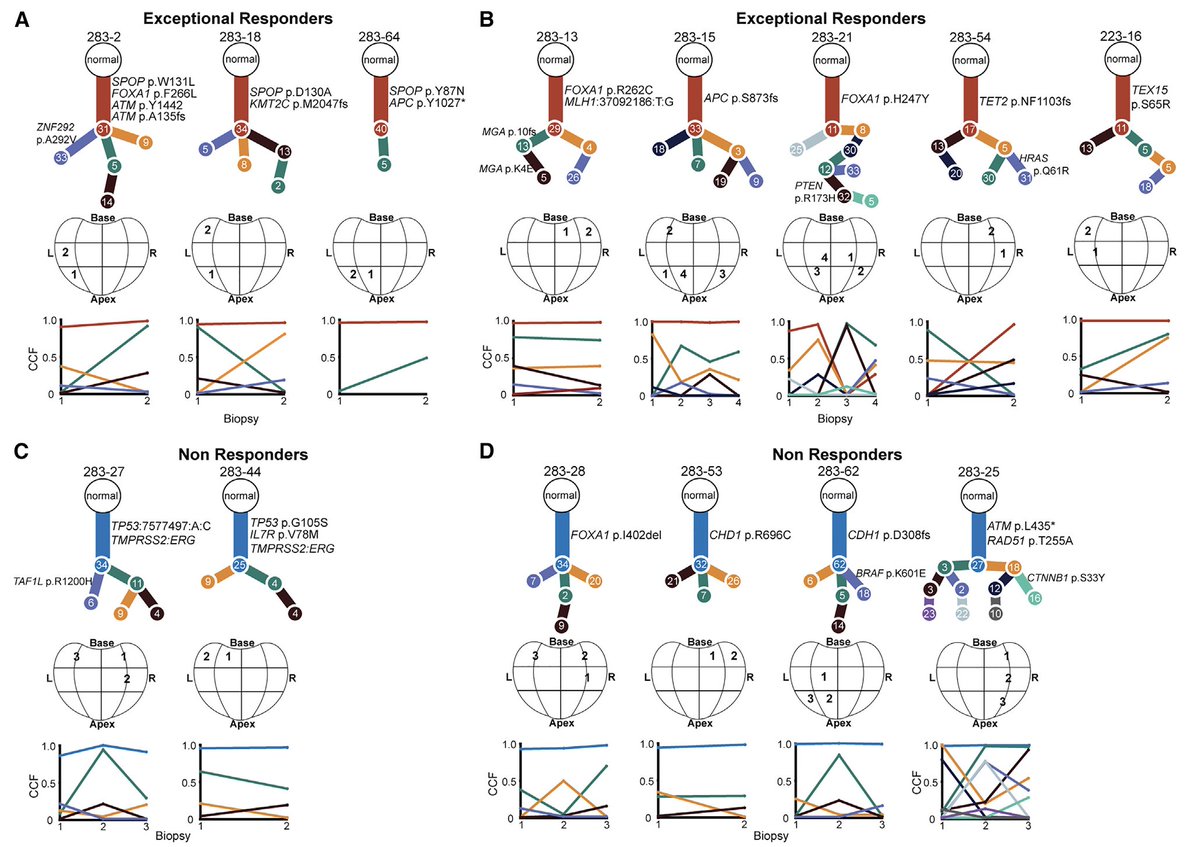
Much more to do...
Hope this study promotes new biology & clinical studies to learn more about underlying molecular states in anti-androgen sensitive tumors
Most importantly, thanks to prostate cancer patients participating in trials & enabling this type of research! [fin]
Hope this study promotes new biology & clinical studies to learn more about underlying molecular states in anti-androgen sensitive tumors
Most importantly, thanks to prostate cancer patients participating in trials & enabling this type of research! [fin]
• • •
Missing some Tweet in this thread? You can try to
force a refresh