We are excited to share our recent work uncovering specialized genome structures in the brain, online today in @Nature:
rdcu.be/cBvwz
@apombo1 @BIMSB_MDC @MDC_Berlin @helmholtz_de @4DNucleome @NeuroCureBerlin
↓ Short thread below ↓
1/7
rdcu.be/cBvwz
@apombo1 @BIMSB_MDC @MDC_Berlin @helmholtz_de @4DNucleome @NeuroCureBerlin
↓ Short thread below ↓
1/7
Led by @WarrenWinick and co-first authors Alexander Kukalev, @IzabelaHC @lunazere9 @domSzabo, this work was an incredible multi-disciplinary effort with 7 international labs:
@GoCasteloBranco @LonnieRWelch Mario Nicodemi, Mark Ungless, Georg Dechant, @teichlab @AltunaAkalin
2/7
@GoCasteloBranco @LonnieRWelch Mario Nicodemi, Mark Ungless, Georg Dechant, @teichlab @AltunaAkalin
2/7
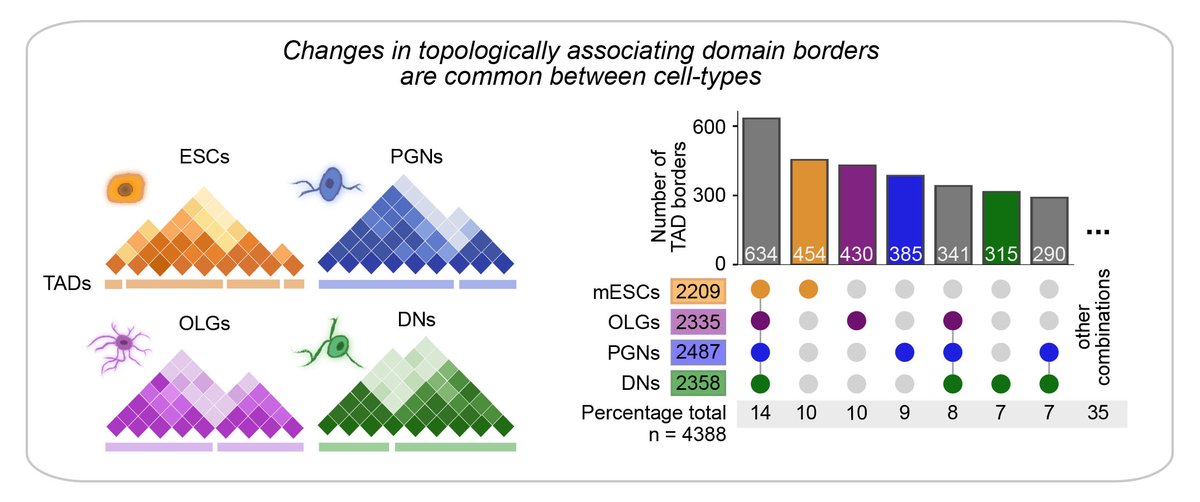
We uncover that specialized brain cells contain highly specific 3D genome structures at multiple genomic scales. For example, only 14% of large-scale TAD borders are common to all cell-types, while cell-type unique borders contain genes relevant for specialized functions.
3/7
3/7
One of the most striking findings was the massive decondensation, or ‘melting’, of long genes when highly expressed or with accessible chromatin. Melting was often accompanied by localization changes, typically towards the interior of the chromosome.
4/7
4/7
Together with @LonnieRWelch and fantastic PhD student Yingnan Zhang, we show that the most differential contacts between neuronal subtypes often contain differentially expressed genes, and are rich with networks of accessible and highly expressed transcription factor motifs.
5/7
5/7
Lastly, we demonstrate that sensory receptor genes form strong clusters of contacts in brain cells. These gene clusters are more strictly found within repressive B compartment regions in brain cells, despite having almost no expression in brain cells or in stem cells.
6/7
6/7

A big thank you to the fantastic reviewers and editor who highly encouraged us to push concepts to the next level.
As a bonus, below is our covert art submission inspired from Dali’s Persistence of Memory, illustrated by lead author @WarrenWinick #SciArt
7/7
As a bonus, below is our covert art submission inspired from Dali’s Persistence of Memory, illustrated by lead author @WarrenWinick #SciArt
7/7

• • •
Missing some Tweet in this thread? You can try to
force a refresh




