
Just as there is notably wider interest in chr1q abnormalities in myeloma now than when our data was published: here again a link to the excellent work chr1q copy number evolution and its clinical impact from James Croft on #UKMRA #MyelomaXI trial nature.com/articles/s4137… 

By investigating new copy number aberrations detected at relapse in 179 matched presentation-relapse tumours from #MyelomaXI, James discovered newly detected 1q aberrations to be most frequent, followed by del13q and del17p. As previous tweet shows, with equal adverse OS. 

DigitalMLPA, the method used to detect these, is sensitive and allowed to look up how many of new 1q relapses had sub-clonal gain(1q) at presentation: answer is some, but not the majority. 

There were other interesting findings, such as that amp1q only in the minority of cases occurred in isolation - most amp1 tumours also carried a high-risk translocation or another high-risk CNA. 
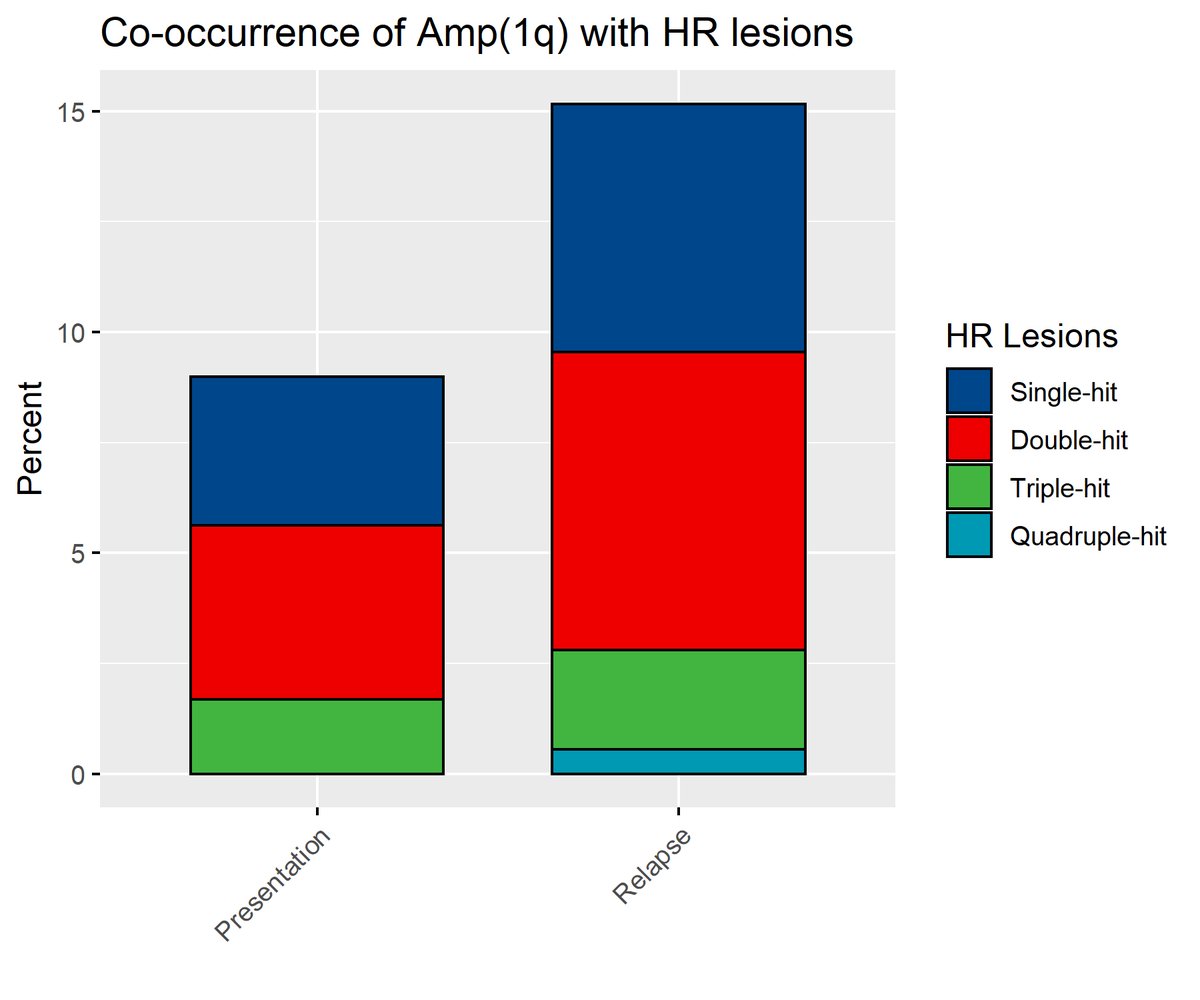
Newly detectable del1p was also associated with inferior OS, as was newly detectable MYC gain. With increasing choice of relapse treatments, repeat genetic profiling should ideally become more widely accessible to myeloma patients (currently not the case in many countries). /Fin 

• • •
Missing some Tweet in this thread? You can try to
force a refresh



