We have trained ESMFold to predict full atomic protein structure directly from language model representations of a single sequence. Accuracy is competitive with AlphaFold on most proteins with order of magnitude faster inference. By @MetaAI Protein Team.
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
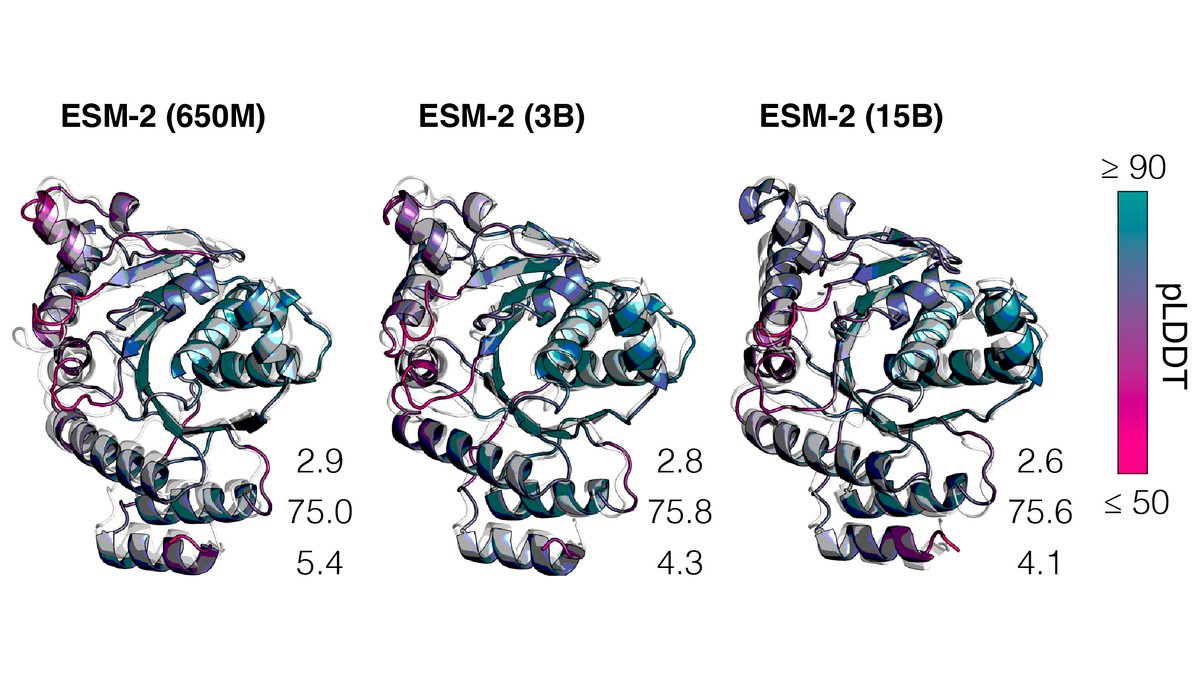
We train ESM2 language models from 8M up to 15B parameters. Improvements in language modeling perplexity and learning of structure continue through 15B. ESM2 at 150M parameters is better than ESM1b at 650M parameters. 

As ESM2 processes a protein sequence, a picture of the protein’s structure materializes in its internal states that enables atomic resolution predictions of the 3D structure, even though the language model was only trained on sequences. 
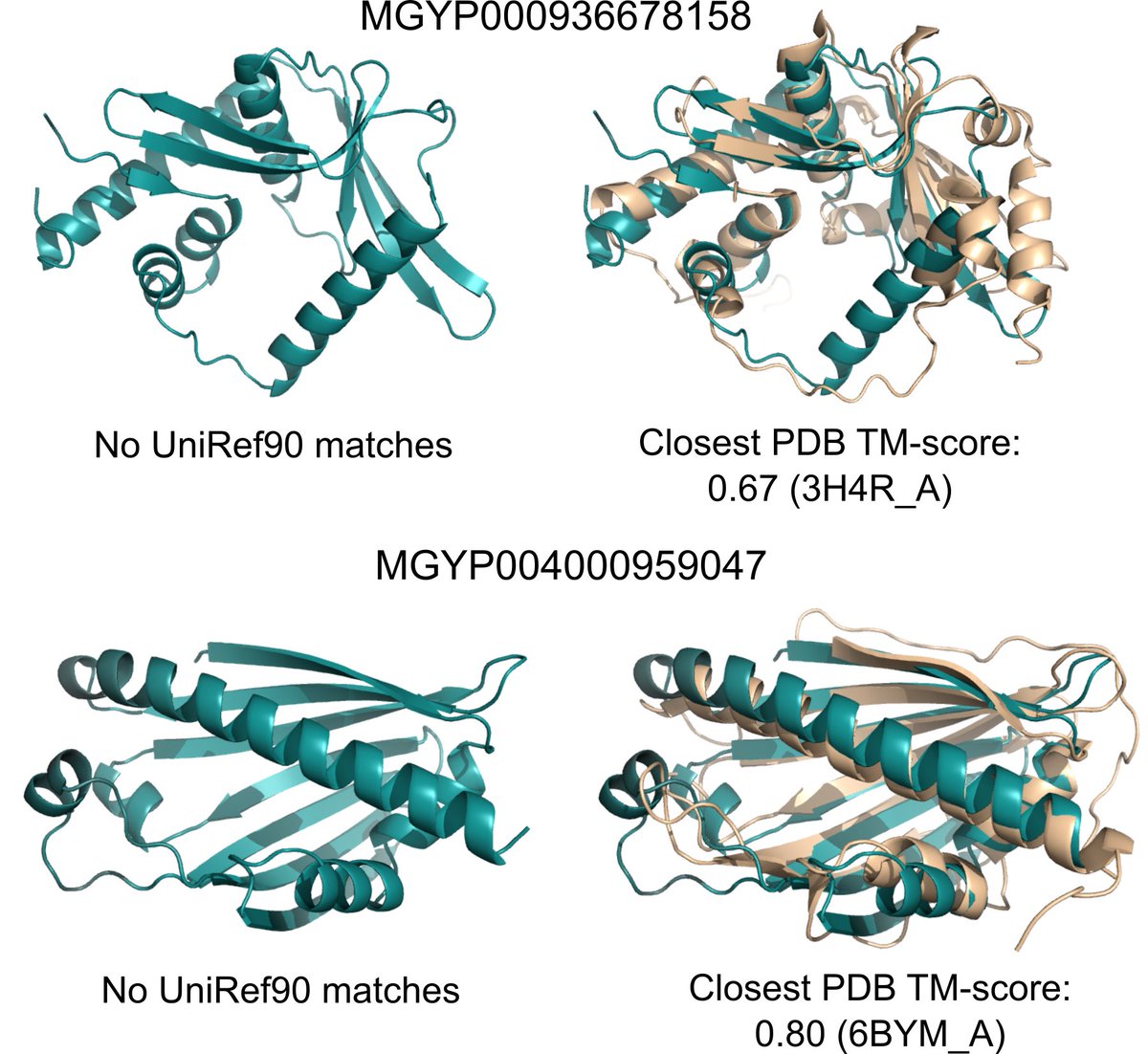
There are billions of protein sequences with unknown structure and function, many from metagenomic sequencing. ESMFold makes it feasible to map this structural space in practical timescales. We were able to fold a random sample of 1M metagenomic sequences in a few hours. 

A large fraction have high confidence and are different from any known experimental structure. Many have sequences without matches in annotated sequence databases. We think ESMFold can help to understand regions of protein space that are distant from existing knowledge. 
Work by the Protein Team at Meta AI FAIR @ebetica @halilakin @proteinrosh @BrianHie @ZhongkaiZhu Wenting Lu @AllanSanCosta Maryam Fazel-Zarandi @TomSercu @salcandido
Building on great open source projects @open_fold @fairseq fairscale foldseek, and many others.
Building on great open source projects @open_fold @fairseq fairscale foldseek, and many others.
• • •
Missing some Tweet in this thread? You can try to
force a refresh






