Just catching up with the literature and noticing that massive research effort by my @KewScience colleagues on the Plant and Fungal Tree of Life Project (PAFTOL) has revealed an Angiosperm353 tree with major differences to the Angiosperm Phylogeny Group (APG IV) tree 🧵(1/9) 
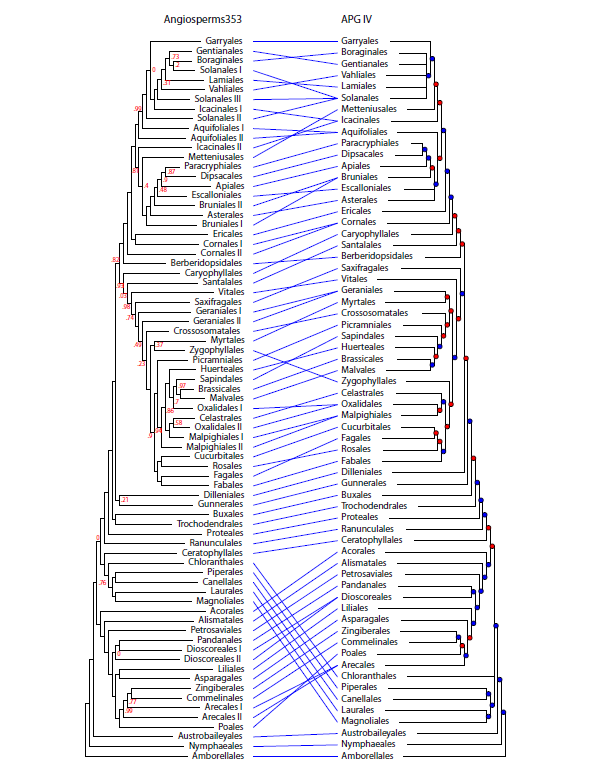
For example, Solanales, Aquifoliales, Bruniales and Icacinales are polyphyletic in the new Angioserm353 tree, and a big polytomy has disappeared (2/9) 
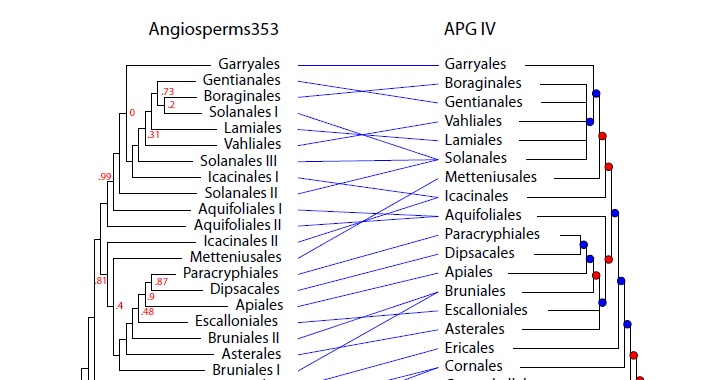
Fagales and Fabales are sister groups in the PAFTOL tree, and Oxalidales and Malpighiales are polyphyletic (3/9) 

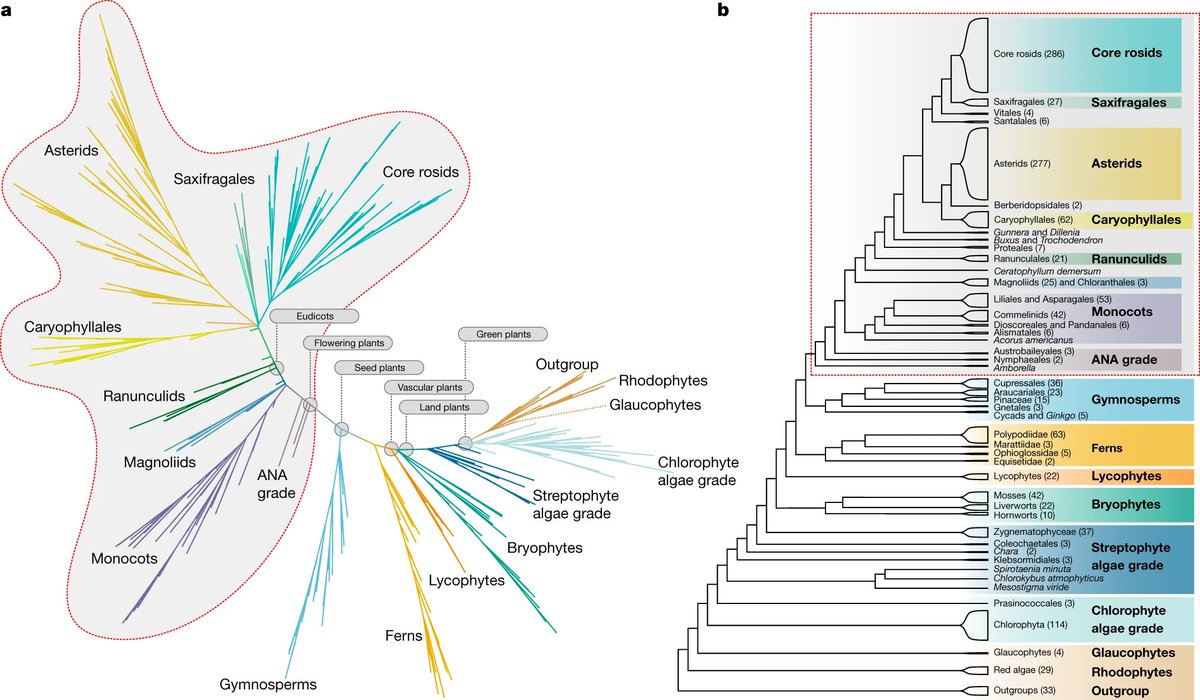
But perhaps the biggest surprise is the placement of the Magnoliids (Magnoliales, Laurales, Cenellales and Piperales) and Chloranthales by PAFTOL. These now appear to be in a clade with the Eudicots! (4/9) 
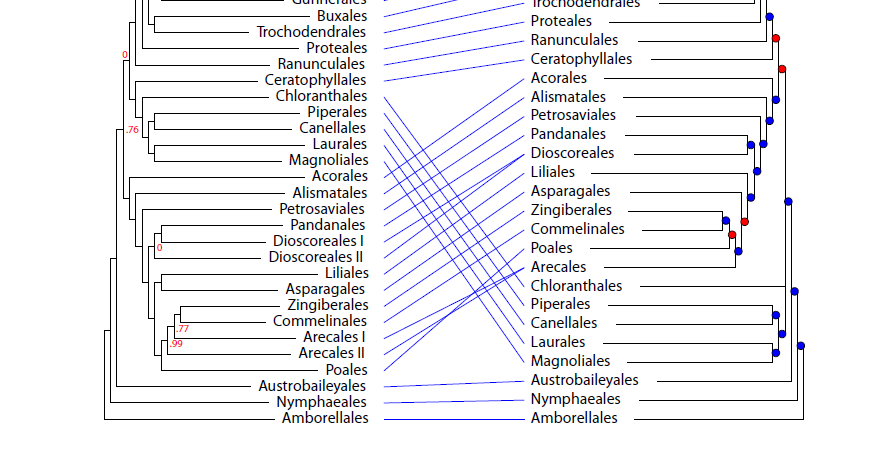
Thus, the Magnoliids diverge after the Monocots in the PAFTOL tree, unlike the placement in this much-used depiction of the APG IV tree (5/9) 
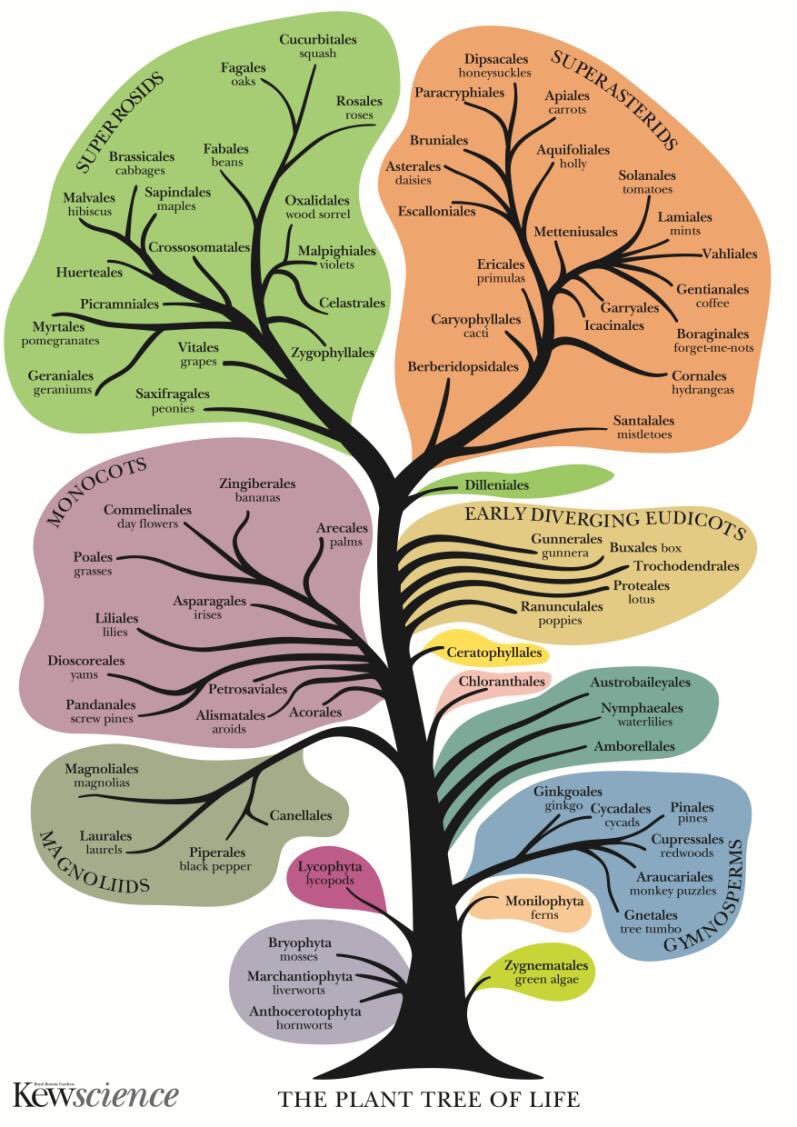
The authors suggest that this is due to lack of resolution in the APG IV tree and "phylogenetic conflict between nuclear & plastid genomes". "It is hardly surprising, then, that a large-scale nuclear analysis presents strongly supported, alternative relationships" (6/9)
They highlight a "conundrum": "these incongruences are visible at the ordinal backbone, but not the family level". If incongruences were due to incomplete lineage sorting and hybridization they we would expect them to be more common at lower taxonomic levels, not higher (7/9)
This is all provisional, and these are preliminary analyses of this vast project, but they could signal a big upheaval for plant taxonomy (in some ways moving back to older groupings) and challenge our current understanding of plant evolution (8/9)
The paper is here. Great work by authors @BillJBaker @DrSDodsworth @FelixForest1 @AlexZuntini @w_eiserhardt @PaulJKersey @IliaLeitch et al @KewScience - and do correct me if anything in the above thread is mistaken (9/9) academic.oup.com/sysbio/article…
I should add that the figure I tweeted is in the supplementary materials on page 17, which can be found here: datadryad.org/stash/download… (10/9)
Note that the 1000 transcriptomes project also gave a similar placement to the monocots and magnoliids & Chloranthales. See nature.com/articles/s4158… (11/9) 
• • •
Missing some Tweet in this thread? You can try to
force a refresh