
What we call (or don't call) a virus plays an important role in how we frame our communications and responses to it. Here is a long thread about how SARS-CoV-2 variants are named.🧵 

What the virus is called has been recognized as important since the early days of the pandemic. Geography-based names were used intentionally by some and avoided as problematic by others because they can promote xenophobia. Think "Wuhan virus" or "China virus".
Even technical names were treated with caution early on. In February 2020, the World Health Organization (@WHO) announced that they would refer to "the virus responsible for COVID-19" or "the COVID-19 virus" rather than "SARS-CoV-2" or "SARS2".
who.int/emergencies/di…
who.int/emergencies/di…

(As you probably know:
COVID-19 = c͟o͟ronav͟i͟rus d͟isease 2019
SARS = s͟evere a͟cute r͟espiratory s͟yndrome
SARS-CoV-2 = SARS coronavirus 2)
COVID-19 = c͟o͟ronav͟i͟rus d͟isease 2019
SARS = s͟evere a͟cute r͟espiratory s͟yndrome
SARS-CoV-2 = SARS coronavirus 2)
Depending on where you live, the original ("wild type") version of SARS-CoV-2 may have caused two major waves of infections/hospitalizations/deaths. Since then, subsequent waves have been caused by the emergence and spread of new variants.
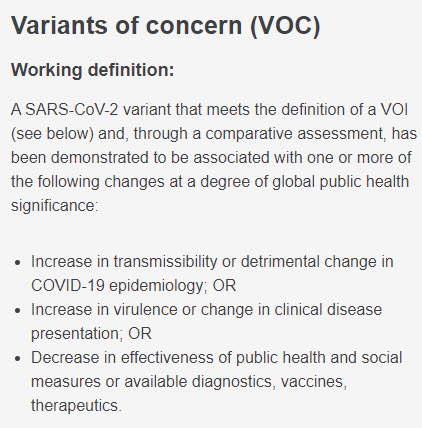
In late May 2021, @WHO announced a system by which new variants could be identified as “variants of interest” (VOIs) or, if they meet additional criteria, “variants of concern” (VOCs).
who.int/news/item/31-0…
who.int/news/item/31-0…

Under this system, VOIs and VOCs are given a label based on the Greek alphabet. To date, @WHO has assigned Greek letter names to eight VOIs (Epsilon, Zeta, Eta, Theta, Iota, Kappa, Lambda, Mu; and five VOCs (Alpha, Beta, Gamma, Delta, Omicron).
who.int/activities/tra…
who.int/activities/tra…
The @WHO decided to skip the letters Nu and Xi between Mu and Omicron in order to avoid confusion with the word “new” and the name of Chinese leader Xi Jinping.
nytimes.com/2021/11/28/wor…
nytimes.com/2021/11/28/wor…
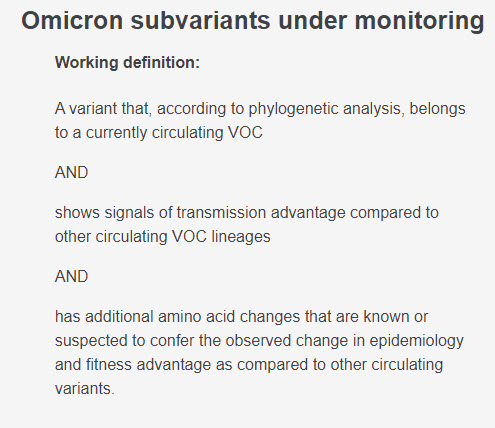
No new VOIs or VOCs have been granted a Greek letter name since the emergence of Omicron in November 2021. All currently circulating variants are considered to be subvariants of Omicron, with @WHO now also recognizing a category dubbed Subvariants Under Monitoring (SUM).
So, how does @WHO decide what counts as a Variant of Interest (VOI), a Variant of Concern (VOC), or a Subvariant Under Monitoring (SUM)?
Here are the criteria they use:
who.int/activities/tra…


Here are the criteria they use:
who.int/activities/tra…

One reason that @WHO has not designated any new Greek letters (the next of which would be Pi, then Rho) is that this system now generally refers to major lineages of SARS-CoV-2 that are not descended from each other.
Omicron is not descended from Delta, and Delta was not a descendant of Alpha. Rather, they share more distant common ancestry, having arisen separately from deeper branches in the SARS-CoV-2 evolutionary tree. 

You can think of the Greek letters as referring to higher-level taxonomic groups. For example, "Omicron" is like "mammals" and Delta is "birds" (though as we will see, Omicron has much more lower-level diversity than previous variants).
All currently circulating variants are members of the large and rapidly evolving Omicron clade. (Clade = an evolutionary group consisting of a common ancestor and all/only its descendants).
That doesn't mean everything is "still just Omicron". Whales and bats are both mammals but are very different biologically. In fact, the different lineages of Omicron variants (think families of mammals, each containing multiple genera and species) are quite divergent.
A few figures from nextstrain.org show just how different Omicron variants (yellow, orange, red) are from each other and from previous variants. The first is an evolutionary tree (in radial format) scaled to divergence (number of mutations). 

The second is a plot of total number of mutations versus sampling date for different variants. The Omicron lineages are above the line, meaning that they are mutating more rapidly than has been typical for the virus. 

The third shows spike protein gene (S1) mutations in particular. Here, Omicron lineages are really evolving at a different pace. 

In the absence of new Greek letter names from @WHO (which, again, are only being applied to higher-level evolutionary groupings at this point), most current discussions of variants refer to PANGO lineage designations. What are those and how do they work?
Phylogenetic Assignment of Named Global Outbreak (PANGO) lineages are similar to formal scientific species names and are assigned according to a suite of detailed evolutionary and epidemiological criteria. You can see these here:
pango.network/the-pango-nome…
pango.network/the-pango-nome…
The short version is that prospective new PANGO lineages are evaluated by a committee based on criteria such as novel evolutionary features, transmissibility, pathogenicity, notable increase in frequency, new movement across regions, etc.
PANGO lineage labels use an alphabetical prefix (proceeding in sequence A, B,... then AA, AB..., etc.; the letters I and O are omitted to avoid confusion with the numbers 1 and 0) and a numerical suffix (e.g., A.1, A.2, A.3, ... B.1, B.2 ... AA.1, AA.2, ... BA.1, BA.2... etc.).
For example, BA.2 has spawned many descendants, as reflected by labels like BA.2.75. Direct descendants of those variants get an additional level in the suffix: BA.2.75.2 is a direct descendant of BA.2.75. 

All of the currently circulating variants are part of the Omicron group, and specifically are members of the BA.2 and BA.5 lineages within Omicron. (Technically, BA.5 and BA.2 are both part of a clade descended from the first BA.2 variant). 

Like wild type, Alpha, Delta, and other variants, the BA.1 lineage of Omicron (of which B.1.1.529 caused the first Omicron wave) is no longer in circulation, having been replaced by descendants of BA.2. 

Some PANGO labels clearly indicate whether they are members of the BA.2 (e.g., BA.2.3.20) or BA.5 lineages (e.g., BA.5.6.2), but what about names like BF.7, BQ.1.1, and XBB?
PANGO labels have a maximum depth of three layers of descendants, and then the label rolls over to a new prefix. For example, BQ.1.1 is an alias for 

A separate system is used to indicate recombinant (hybrid) variants, in this case beginning with X followed by another letter(s) (proceeding in sequencing XA, XB,... then XAA, XAB...) and containing no numerical suffix except for unambiguous descendants (e.g., XB, XB.1).
A third naming system is used by Nextstrain.org (and CoVariants.org), consisting of a numerical prefix representing the year in which the clade was designated followed by a letter given in order (e.g., 20A, 20B... 21A, 21B, 21C).
Nextstrain.org clades represent a small subset of PANGO lineages that meet additional growth or frequency criteria. These are the clades that appear colour-coded in the phylogenies on Nextstrain.org.
nextstrain.org/blog/2022-04-2…
nextstrain.org/blog/2022-04-2…

All of this can get a bit confusing, because any given variant may have four names: @WHO Greek letter, PANGO full lineage, shorter PANGO alias, and Nextstrain clade. 

In terms of formal names, we have both high level (Greek letters) and variant by variant technical names (PANGO lineages). Again, this is similar to having "mammals" as the broad category and scientific names for each variant.
This is fine for technical discussions, but it can become cumbersome when attempting to communicate more broadly. Consider the multiple variants being watched right now (early Oct. 2022): 

Formal names are important in biology, but in non-technical communication about species that we encounter most often or are otherwise of interest to us (e.g., dangerous, cuddly, delicious) we usually make use of common names. 

Unless you're a zoologist, you probably recognize the species listed in the previous table much more easily by their common names. 

In a similar way, there is some interest in having a system for assigning "common names" or "nicknames" to SARS-CoV-2 variants that are of the most interest. The first was "Centaurus" for BA.2.75, proposed by @xabitron1.
Other names based on Greek mythological creatures have been suggested, though this is really a stop-gap until a proper system can be put in place for assigning common names for variants. 

* B.Q.1.1 is an alias for B.1.1.529.5.3.1.1.1.1.1.1
There are some important differences now versus in the past with regard to variant evolution and diversity. As noted, @WHO Greek letters refer to very distinct/distantly related lineages, and there haven't been any of those detected since Omicron.
It seems that major new lineages may evolve within individual hosts with persistent infections, specifically people who are immunosuppressed or immunocompromised. This is all the more reason to protect the vulnerable and prevent more people from experiencing immune suppression.
nejm.org/doi/full/10.10…
nejm.org/doi/full/10.10…
science.org/content/articl…
nature.com/articles/s4159…
nature.com/articles/s4146…
scientificamerican.com/article/covid-…
nejm.org/doi/full/10.10…
science.org/content/articl…
nature.com/articles/s4159…
nature.com/articles/s4146…
scientificamerican.com/article/covid-…
What we're seeing with the rapid evolution of closely-related Omicron lineages is more likely the result of evolution at the human host population level rather than within individual hosts.
Unlike past waves, which were typically caused by the replacement of one variant by another, we now have multiple variants circulating at the same time, each having evolved under natural selection for immune escape. 


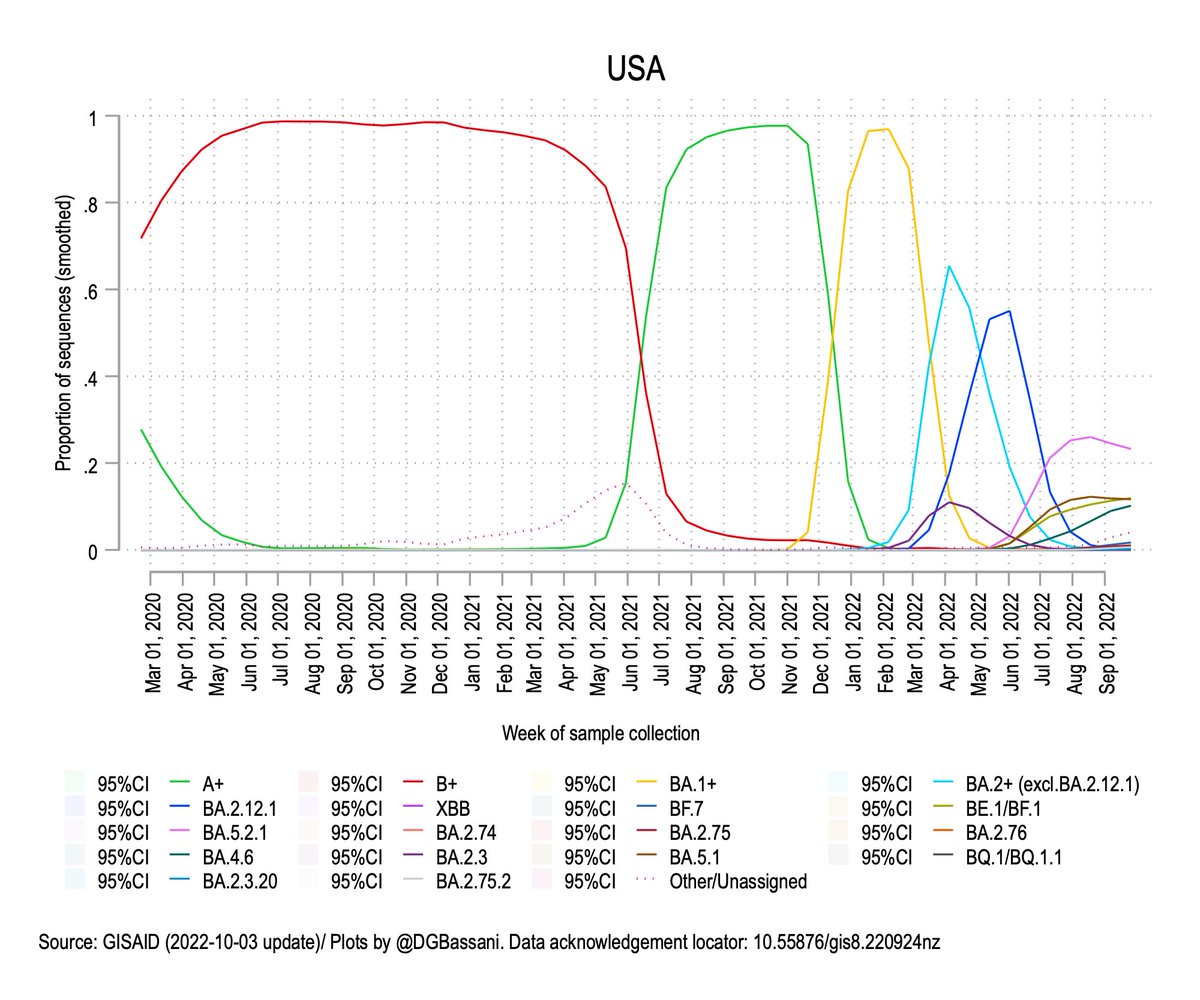
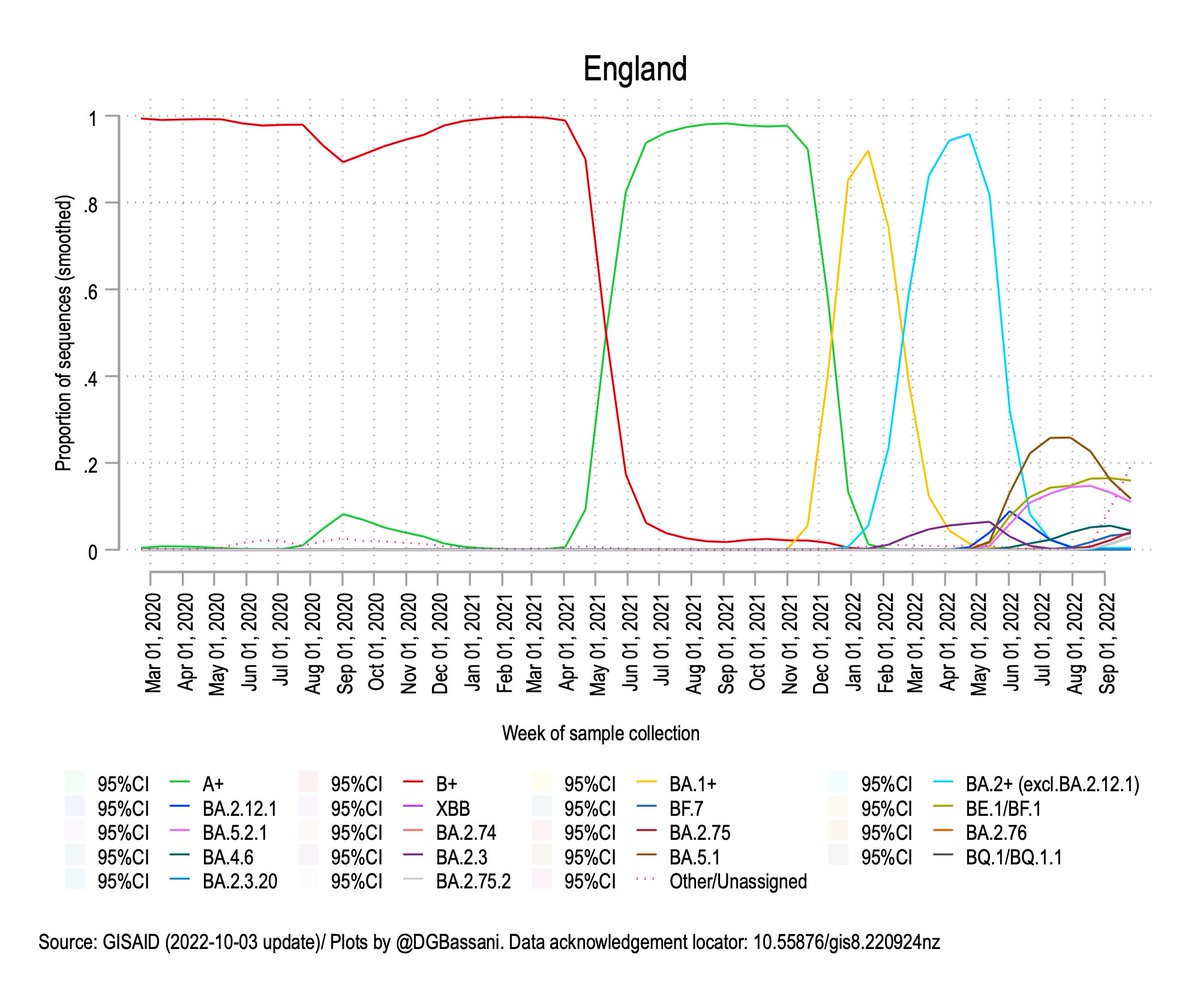

So what does this all mean? A few things:
* We need to continue paying attention to variant evolution. Having ways to communicate effectively about this will be important.
* It's not "all just Omicron" any more than whales and bats are "both just mammals".
* We need to continue paying attention to variant evolution. Having ways to communicate effectively about this will be important.
* It's not "all just Omicron" any more than whales and bats are "both just mammals".
* Having multiple immune-escaping variants at once is new. We don't know what the impacts will be, but there are reasons to take this seriously.
science.org/content/articl…
science.org/content/articl…
* We could still end up with a very different lineage of variant (i.e., not Omicron but Pi or Rho). It could be out there evolving in someone with a persistent infection as we speak.
* "Robust hybrid immunity" is a pipedream if immune-escaping variants keep evolving rapidly.
* "Robust hybrid immunity" is a pipedream if immune-escaping variants keep evolving rapidly.
* More transmission means more viral copies out there replicating, which means more mutations added to the viral gene pool. Large populations sizes of viruses also means more powerful natural selection. That is, unmitigated transmission = more variants.
If you want to stay up to date on new variants, I recommend following everyone on this list:
twitter.com/i/lists/156947…
twitter.com/i/lists/156947…
You can also access sources of information directly. Here's a thread with a list of resources:
https://twitter.com/TRyanGregory/status/1569478277160660993?s=20&t=niEmeu6r4M2OZ5JR8CVrZQ
• • •
Missing some Tweet in this thread? You can try to
force a refresh















