"Best practices for single-cell histone modification analysis" w/ @VallotCeline @jeanphi_vert and @PPrompsy.
We identify how to analyze scCUT&Tag, from matrix construction, QC, #cells, up to dimension reduction.
paper: biorxiv.org/content/10.110…
code: github.com/vallotlab/benc…
We identify how to analyze scCUT&Tag, from matrix construction, QC, #cells, up to dimension reduction.
paper: biorxiv.org/content/10.110…
code: github.com/vallotlab/benc…

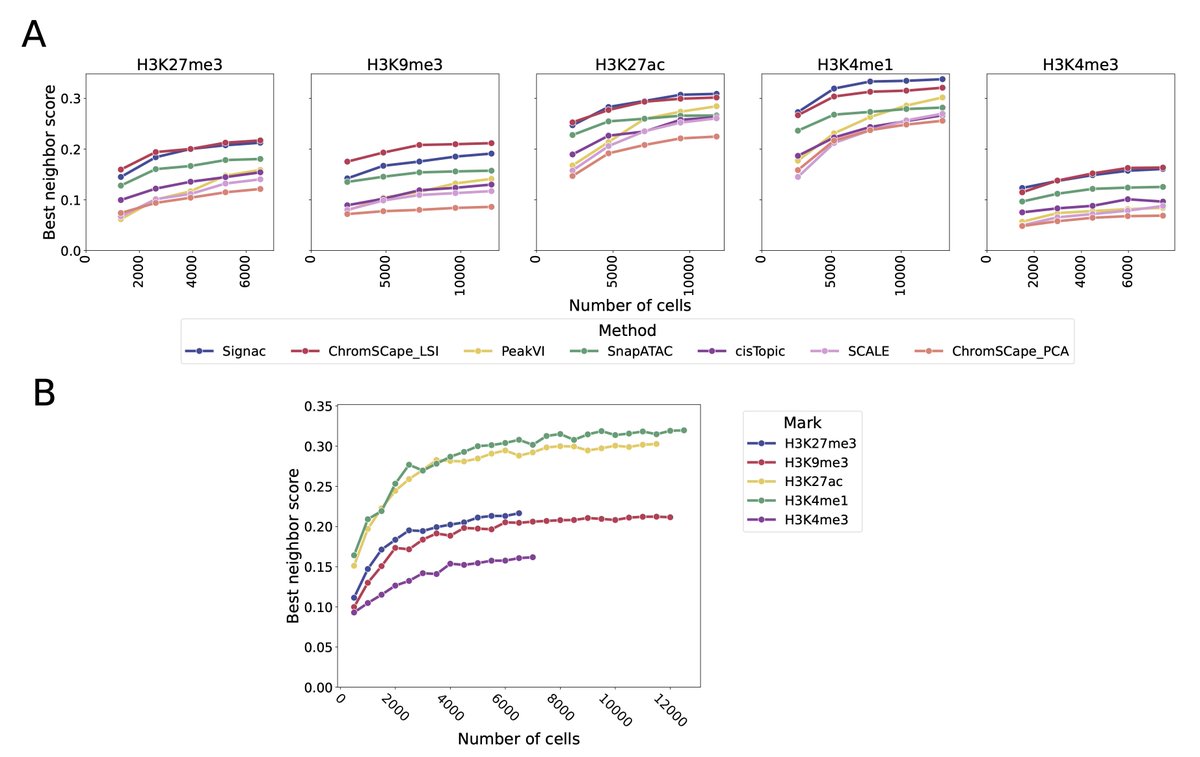
@VallotCeline @jeanphi_vert @PPrompsy We use multiomics assays to evaluate the quality of the representation in a completely unsupervised fashion, by using the kNN-AUC between the two modalities (see openproblems.bio/benchmarks/mul…)
We further look at the 5 most common histone marks on human PBMC and mouse brain data.
We further look at the 5 most common histone marks on human PBMC and mouse brain data.

@VallotCeline @jeanphi_vert @PPrompsy We show that similarly to scATAC-seq, LSI-based methods such as Signac and ChromSCape tend to outperform their competitors.
We further show that enhancing marks (H3K4me1 and H3K27ac) have extremely high agreement with expression data.
We further show that enhancing marks (H3K4me1 and H3K27ac) have extremely high agreement with expression data.

@VallotCeline @jeanphi_vert @PPrompsy We investigate the role of matrix construction on the quality of the representation, and find that a proper contraction can increase the performances by up to 80%.
We also show that using peaks called on pseudo bulk, or a GeneTSS annotation is worse than bins of the right size
We also show that using peaks called on pseudo bulk, or a GeneTSS annotation is worse than bins of the right size

@VallotCeline @jeanphi_vert @PPrompsy We identify that feature selection methods based on coverage or HVG are always detrimental to the quality of the representation. 

@VallotCeline @jeanphi_vert @PPrompsy We also identify that while increasing the number of cells in an experiment is always beneficial, its effect tends to saturate around the 6.000 cells mark.
The only method not saturating being the VAE-based PeakVI, but it still fails to outperform LSI in the 12.000 cells regime
The only method not saturating being the VAE-based PeakVI, but it still fails to outperform LSI in the 12.000 cells regime
@VallotCeline @jeanphi_vert @PPrompsy This work was made possible by my employer at the time @GoogleAI, which allowed us to run an extremely large amount of computations.
The expertise from @institut_curie allowed us to go beyond simple machine learning, and answer questions relevant to practicing biologists.
The expertise from @institut_curie allowed us to go beyond simple machine learning, and answer questions relevant to practicing biologists.
• • •
Missing some Tweet in this thread? You can try to
force a refresh




