
On phylogenetics (evolutionary trees), monophyly (clades), and paraphyly. And SARS-CoV-2 variant evolution, of course. 🧵
(All of these ideas are covered in detail in this open access articel: evolution-outreach.biomedcentral.com/articles/10.10…).
(All of these ideas are covered in detail in this open access articel: evolution-outreach.biomedcentral.com/articles/10.10…).
First, some obligatory reference to etymology and Greek word origins.
Phylogenetics / phylogeny: Greek phûlon ("tribe, clan, race") + genetikós ("origin, source, birth").
Monophyletic / monophyly: Greek moûnos ("one") + phûlon ("tribe, clan, race").
Phylogenetics / phylogeny: Greek phûlon ("tribe, clan, race") + genetikós ("origin, source, birth").
Monophyletic / monophyly: Greek moûnos ("one") + phûlon ("tribe, clan, race").
Paraphyletic / paraphyly: Greek pará ("beside, near") + phûlon ("tribe, clan, race").
Clade: Greek kládos (“shoot, branch”).
Clade: Greek kládos (“shoot, branch”).
You've likely seen the term "phylum" as well for high level taxononomic groups below the level of kingdom and above the level of class (e.g., chordates, arthropods, molluscs, etc.) -- same origin.
I'm not going to get into a lot of detail about reading trees here, but I do want to talk about what we mean by "clade", "monophyly", and "paraphyly", and why that is relevant to understanding SARS-CoV-2 variant evolution.
The term "monophyly" refers to a grouping of lineages that includes the common ancestor, all of its descendants (not missing any), and no lineages that arose outside that group.
Monophyletic groups -- and only monophyletic groups -- are called "clades".
Clades are nested inside larger clades as you extend back in time / to deeper nodes in the tree.
Clades are nested inside larger clades as you extend back in time / to deeper nodes in the tree.

If a group includes the ancestor and *some* (but not all) of its descendants, it is not monophyletic and thus not a clade. Rather, it is paraphyletic.
Image source: online.ucpress.edu/abt/article-ab…
Image source: online.ucpress.edu/abt/article-ab…

A quick an easy way to know if you have a clade is to imagine cutting the tree at a particular branch. If you make one cut and grab the cut branch, everything you're holding is a clade. If you have to cut two branches to make the group, it's not a clade.
Another way of saying this is that if you have two groups with names, and one is nested within the other phylogenetically, then only the one nested within the other is a clade. The one in which that clade is nested is paraphyletic. (This will be clear in the next tweets).
So, let's think about "birds" and "reptiles".
Birds are all descended from a common ancestor, and the named group "birds" includes all of the descendants and no lineages from outside that group. So "birds" is monophyletic, i.e., a clade.
Birds are all descended from a common ancestor, and the named group "birds" includes all of the descendants and no lineages from outside that group. So "birds" is monophyletic, i.e., a clade.
But "birds" are nested within the group we commonly refer to as "reptiles". Put another way, some "reptiles" (specifically crocodilians) are more closely related to birds than they are to other "reptiles". Here's the figure from Baum and Offner (2008) once again: 

To make a group named "reptiles", you have to group all the "reptiles" (lizards, snakes, turtles, crocodiles, etc.) and cut out birds. So one descendant lineage is left out, and "reptiles" is not a clade.
There are lots of familiar groups with names that are paraphyletic: "invertebrates", "fishes", and "monkeys" are all paraphyletic. 

In terms of scientific classification, there is generally a view that taxonomic names should only be given to clades. However, in broader discussions, it can be useful to refer to groups by a name even if they're paraphyletic. So don't worry about saying "reptile" or "fish".
Ok, so now on to SARS-CoV-2 variants. These also evolve through a branching process involving nested groupings of ancestors, descendants, and more or less closely related lineages. In general, we refer to clades and give those labels.
Image: covariants.org
Image: covariants.org

Most recently, there has been extensive branching of new lineages within the large Omicron clade.
Images: nextstrain.org and @dfocosi

Images: nextstrain.org and @dfocosi


With Omicron, we've have several waves this year in various places in the world, each (so far) caused by one variant. BA.1, then BA.2, then BA.5.
BA.1 is no longer circulating. What we've been talking about with new variants are all members of the BA.2 and BA.5 lineages.
BA.1 is no longer circulating. What we've been talking about with new variants are all members of the BA.2 and BA.5 lineages.
The evolution of Omicron lineages is not always simple. For one thing, there are notable examples of hybridization across lineages, most notably XBB which is a recombinant between BJ.1 x BM.1.1.1. 
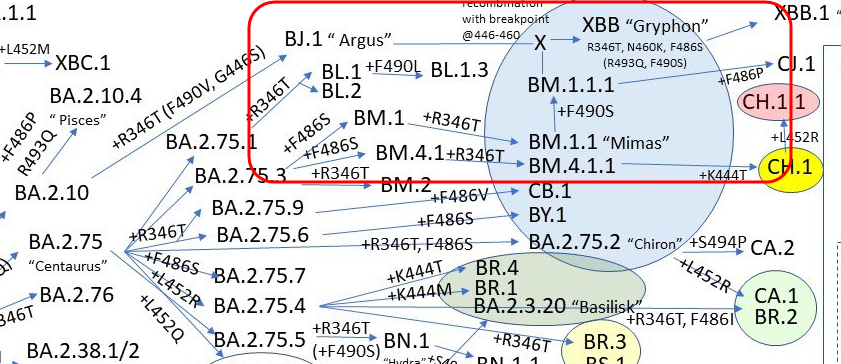
Let's look more closely at the Omicron clade (as a group "Omicron" is monophyletic). First, we see a major split between BA.1 and BA.2 lineages.
(Both Pango [e.g., BA.1] and Nextstrain [e.g., 21K] labels are indicated).
Image: covariants.org
(Both Pango [e.g., BA.1] and Nextstrain [e.g., 21K] labels are indicated).
Image: covariants.org
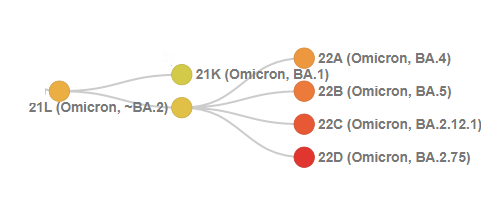
Within BA.2 is where it gets a bit more complicated. It's quite clear that BA.4 and BA.5 are each clades, and that together they form a clade. 

However, the BA.4+BA.5 clade is itself descended from BA.2, and is nested within BA.2. We also have the Nextstrain groups 22C and 22D nested within 21L. 

It is important to bear in mind that the order of branching (topology) shown closer to the tips (terminal nodes) on the tree can be challenging to resolve. The interpretation above is based on assuming that there is no clear split between BA.4+BA.5 and all the other "BA.2"s.
In addition, BA.2 being paraphyletic is contingent on the ancestor of (BA.2+(BA.4+BA.5)) being called BA.2. This is what makes BA.4+BA.5 nested within BA.2.
If the ancestor of (BA.2+(BA.4+BA.5) had not been BA.2, and there had been clear split between BA.4+BA.5 and BA.2, and it would be two clades, like with BA.1 and BA.2 or BA.4 and BA.5.
In the end, it doesn't really matter too much except that we need to be clear to distinguish the ancestral BA.2 that caused its own wave from the descendant lineages still called BA.2 (including BM, etc.) and the other descendant lineages now called BA.5 (including BQ, etc.).
I did manage to make you read a whole thread on evolutionary trees. So that's a win.
I should have linked to this thread on how the names work. One of the complexities is that BA.2, BA.5, BQ.1.1, etc., are aliases that keep the Pango lineages from getting too long.
Original Omicron = B.1.1.529
BA.1 = B.1.1.529.1
BA.2 = B.1.1.529.2
BQ.1 = B.1.1.529.5.3.1.1.1.1.1
Original Omicron = B.1.1.529
BA.1 = B.1.1.529.1
BA.2 = B.1.1.529.2
BQ.1 = B.1.1.529.5.3.1.1.1.1.1
• • •
Missing some Tweet in this thread? You can try to
force a refresh











