Fantastic talk by @kapoormanav on the genetic architecture of South Asian Indian population based on analysis of more than 15,000 exomes from Rajasthan and Maharashtra states of India. #ASHG22
https://twitter.com/doctorveera/status/1586021752307695616?s=20&t=yyRkvOwSlz0K67QYTEbo-Q
India is one of the most diverse country in the world with 22 official spoken languages, ~3000 castes, 25,000 sub castes and a population size of 1.4 billion. And yet India is one of the most underrepresented countries in the genetic studies.
The extreme diversity in terms language, caste and culture makes Indian population so unique as these factors have sculpted the genetic architecture of Indians over hundreds to thousands of years.
This fascinating fact can be readily appreciated when we cluster the Indian individuals based on their genetic data, which Manav showed in beautiful UMAP plots based on first 10 PCs.
Individuals cluster together based on language they speak more than where they live. Within each language clusters, individuals cluster based on their caste and sub-caste.
https://twitter.com/doctorveera/status/1355474125884633089?s=20&t=8z7cPvBGJe0uxKYRKmNBRg
This is because individuals marry only within a caste and those who marry outside the caste become social outcast and sometimes face grave consequences.
https://twitter.com/doctorveera/status/1313432896842141696?s=20&t=8z7cPvBGJe0uxKYRKmNBRg
Such cultural barriers to gene flow gave birth to thousands of "isolated" populations, similar to geographical barriers resulting in isolated populations (e.g. Northern Isles)
Note, sociocultural barriers are more powerful than geographical barriers to gene flow. This results in founder effects that are stronger than what we've seen in well studied founder populations like Finns and Ashkenazi Jews
https://twitter.com/doctorveera/status/1469874004274008064?s=20&t=8z7cPvBGJe0uxKYRKmNBRg
In the current larger dataset, Manav was able recapitulate the strong founder effects in terms of high within community IBD scores for many caste communities.
Such strong founder effects seem to have caused fascinating consequences on the admixture proportions of the present day Indians.
Manav modeled the admixture of the study participants based on three major ancestries--ancestral South Indians (ASI, Indo-Dravidians), ancestral north Indians (ANI, Indo-Aryans) and Europeans (EUR)
Moving from North to South axis, as expected many communities showed linear increase in ASI and decrease in ANI and EUR. But some communities stood out showing predominantly ANI (Jain) or predominantly ASI (Koli)
This suggests that such communities have experienced strong barriers to gene flow due to strong endogamy practices, which was also reflected in the high IBD scores in those communities. That was fascinating!
Another area that has been poorly explored in Indian context is the positive selection. Humans who settled over this large country many thousand years ago over multiple migrations for sure faced many challenges like food, pathogen etc. and underwent adaptation.
We know little about those such positive selections. Analysing the genetic data of the 15k samples, Manav identified 24 loci across the genome with strong signs of positive selection. Some are known, e.g. LCT, HLA etc.
Some are novel. Manav highlights a locus in chr 4 that contains gene OTOP1 that encoded a zinc sensitive channel protein that plays an important role in inner ear balance. Variants within this positively selected region strongly associate with vertigo.
As a South Asian Indian who has suffered from motion sickness since childhood and diagnosed with vertigo in adulthood, I am particularly fascinated by this finding.
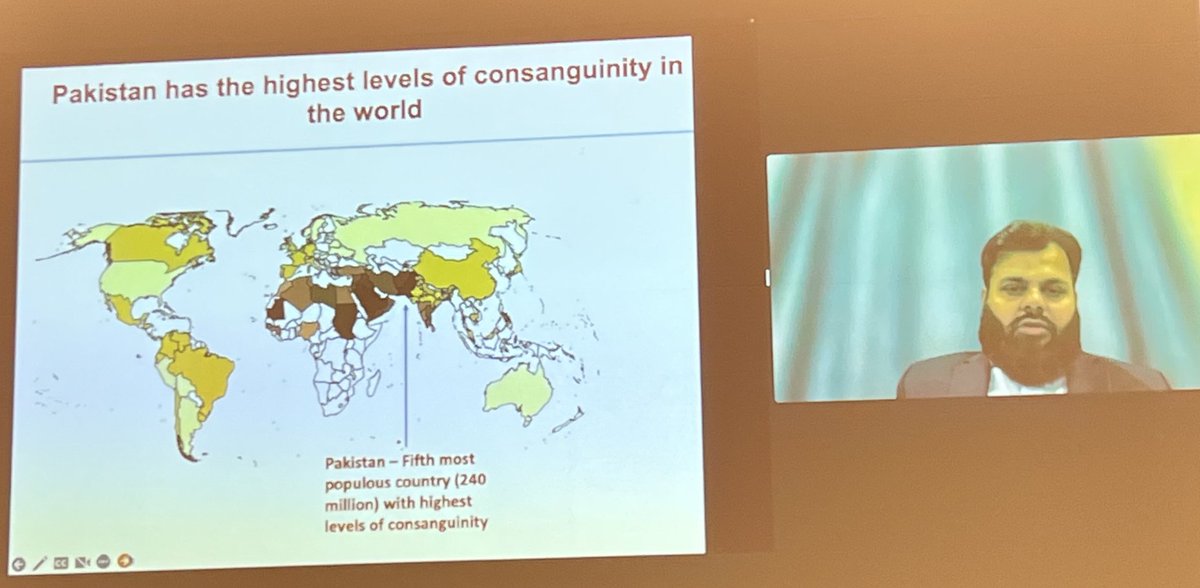
Next, the most important topic: human knockouts. Similar to Pakistan, consanguineous marriages are common in India, particularly within certain caste communities and this result in high frequency of homozygous loss of function mutations.
Manav showed that the 15k Indian samples had 5 times more number of homozygous pLOFs compared to sample size matched European dataset. Many of these pLOFs were never seen before.
Manav highlighted a splice donor variant in PLA2G7 that encodes an enzyme that degrades platelet activating factor. this variant is seen 0.5-1% of the Indians but absent elsewhere in the world.
This variant increases the risk for cardiovascular disease. Individuals carrying this variant are at more than five times odds of having undergone coronary bypass surgery. It seems like there might be hundreds of thousands human knockouts for PLA2G7 roaming around in India.
The initial results that Manav presented look amazing and with increasing sample sizes these results will get even better. Watch this space for future updates.
I noticed that some good geneticists whom I respect made insensitive comments on our work. As an Indian who has lived and seen bad side of the caste system, I want to emphasize strongly that caste is extremely important factor when it comes to genetics in India.
This knowledge is important for both making genetic discoveries, to diagnose diseases and most importantly, for educating the future generations on the bad effects of endogamous practices.
• • •
Missing some Tweet in this thread? You can try to
force a refresh