Are you interested in performing splice-aware quantification of your #scrnaseq data, obtaining unspliced, spliced, and ambiguous UMI counts quickly & in <3GB of RAM? If so, check out the new manuscript by @DongzeHe, @CSoneson and me on #bioRxiv bit.ly/3vJr0Ji. 1/🧵
Understanding the origin of sequencing reads — the molecules from which they arise, the "gene" with which those molecules are associated, and the splicing status of those molecules — is a key task in single-cell RNA-seq quantification.
The short, (effectively) single-end nature of the reads used in popular technologies leads to situations in which it can be difficult or impossible to predict if a read was sequenced from an unspliced (nascent) or spliced (mature) RNA; these reads are designated as "ambiguous."
It turns out, however, that there are multiple ways in which one can define "ambiguous" reads. Prevailing convention considers UMIs associated with reads arising purely from exons as "spliced", so long a there is no contradictory evidence.
An alternative approach is to be more conservative, and to consider reads arising from within exons to simply be "ambiguous". This is the proposal made in a recent preprint (bit.ly/3GjCPuI), and the motivation is intuitive. But what is the effect of this choice?
One immediate effect is that a large fraction of reads in a typical sample now fall into this ambiguous category (see our Table 1). In fact, these numbers are ~5x higher than if one adopts the "traditional" allocation rules. 

While this convention can avoid classifying exonic reads arising from unspliced molecules as spliced, it also doesn't recognize that exonic reads are not equally likely to arise from spliced and unspliced molecules (Fig. 2; caveat, from a vastly simplified simulation). 

Since assignments made by alevin-fry are based on how the reference transcriptome is constructed, simply changing from a splici reference to a spliceu reference (spliced transcripts + one nascent transcript per gene) results, immediately, in the more conservative assignment rule. 

Yet, most downstream analyses have no specific mechanism for dealing with ambiguous reads. So, these reads must either be discarded, or assigned some definitive status. Discarding them would throw away too much information, so a definitive “classification” is typically made.
It turns out that if you choose to classify all ambiguous reads as spliced in scRNA-seq, or unspliced in snRNA-seq (a choice certainly worthy of further investigation), then the more conservative assignment rules produce results that are quite similar to the existing conventions. 

There are differences (and methods adopting the same classification rules tend to be more concordant, as one would expect), but broadly, the resulting quantifications are quite similar. 

Unfortunately, we were not able to straightforwardly reproduce results consistent with previous claims to the contrary, but that may have been due to an annotation error (see Section 2.2 of the manuscript). 

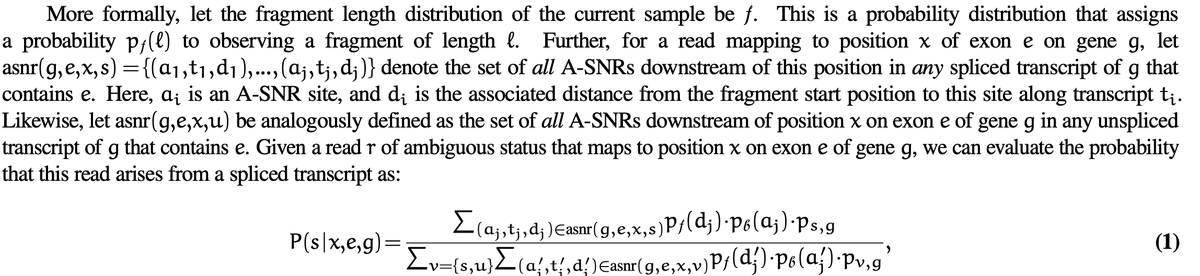
So, how best to assign splicing status still seems to be far from a solved problem! Compared to the relatively simple assignment rules currently used, one enticing direction is to build a more informed probabilistic model that incorporates additional evidence.
One valuable piece of evidence that is not currently used is the location of the likely priming site of the "technical read". This could be incorporated into the evaluation of splicing status. We make some suggestions to this effect in the paper. 
Given that important aspects of this question remain to be resolved, we contend that quantification methods should categorize the splicing status of UMIs according to a clearly-defined set of rules & propagate all UMIs, tagged with their splicing status, to downstream tools.
Rather than deciding what to count and what to discard during mapping and quantification, the relevant information can be propagated to the count matrix so that downstream tools can make informed decisions based on their models and use cases.
Of course, this requires mapping & quantification be performed against a single index containing both spliced & unspliced (nascent) transcripts. To this end, we suggest the piscem index (introduced at #biodata22), which can map against this enhanced transcriptome in <3GB of RAM!
Piscem github.com/COMBINE-lab/pi… can be used directly upstream of alevin-fry as a replacement for the salmon mapper (which uses the pufferfish index). It unlocks many opportunities for indexing large sequence collections beyond scRNA-seq; stay tuned for a more thorough description.
In conclusion, this topic definitely deserves more attention (dare I say "more new algorithms")! In the meantime, it may be best to maximize the information you keep. Luckily, that's easy to do! 19/19
Link back to the top of the thread —
https://twitter.com/nomad421/status/1611352082686025728?s=20&t=1vsn6i_4-XeNFKCYVoSBvg. 20/19
• • •
Missing some Tweet in this thread? You can try to
force a refresh




