
Five papers have been accepted to #ICLR2013 in my group (including one oral presentation), covering topics from combining pretrained LMs with GNNs, deep generative models and pretraining methods for drug discovery.
1) An oral presentation. We proposed an effective and efficient method for combining pretrained LLMs and GNNs on large-scale text-attributed graphs via variational EM. The first place on 3 tasks of node property prediction on OGB leaderboards.
Paper: openreview.net/pdf?id=q0nmYci….
Paper: openreview.net/pdf?id=q0nmYci….

2) An end-to-end diffusion model for protein sequence and structure co-design, which iteratively refines sequences and structures through a denoising network.
Paper: openreview.net/pdf?id=pRCMXcf…
Paper: openreview.net/pdf?id=pRCMXcf…
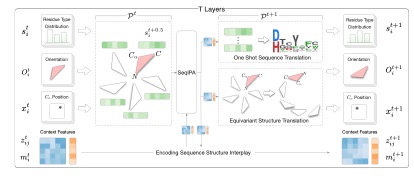
3) An encoder-decoder framework for protein-ligand docking. An encoder is used to encode the representations of protein 3D structure and molecular graph and their interactions, and a diffusion network for predicting the complex structure.
Paper: openreview.net/pdf?id=sO1QiAf…
Paper: openreview.net/pdf?id=sO1QiAf…

4) We pretrained a geometry protein encoder based on the experimental structures in PDB and the predicted structures by AlphaFold 2. It outperforms the sequence-based pretraining methods with protein LMs.
Paper: openreview.net/pdf?id=to3qCB3…
Paper: openreview.net/pdf?id=to3qCB3…

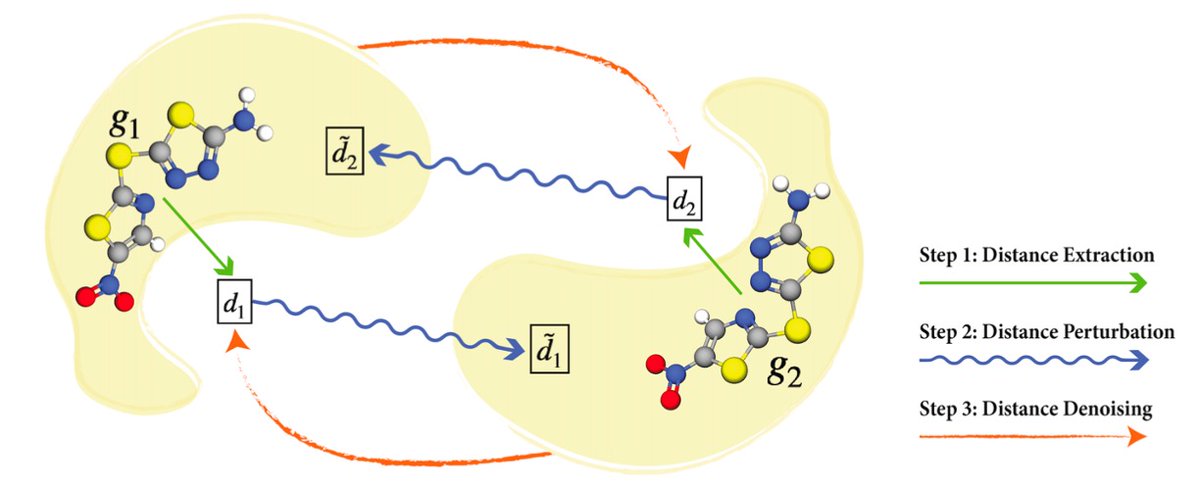
5) We proposed a SE(3)-invariant denoising score matching approach for pretraining molecular geometry representation. State-of-the-art performance on existing molecular pretraining benchmarks. 
Oops, should be #ICLR2023
• • •
Missing some Tweet in this thread? You can try to
force a refresh




