I wanted to highlight this pre-print by David Ho’s group on the neutralizing antibody response to new (XBB.1.5-based) COVID vaccine booster, as it illustrates some points related to paradigm of updating SARS-CoV-2 vaccines to keep pace w viral evolution.
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
Recall original COVID vaccines worked very well against early SARS-CoV-2 strains
Unfortunately, virus has been evolving, so antibodies elicited by that vaccine don’t neutralize newer viral variants very well
(Other human CoVs also evolve same way: ) journals.plos.org/plospathogens/…

Unfortunately, virus has been evolving, so antibodies elicited by that vaccine don’t neutralize newer viral variants very well
(Other human CoVs also evolve same way: ) journals.plos.org/plospathogens/…

So in fall 2022, new booster was made that mixed new (at time) BA.5 variant & original strain. Hope was to boost neutralization of new variants.
Unfortunately, only sort of worked. Titers did go up, but not a relatively greater increase for new variants.
Unfortunately, only sort of worked. Titers did go up, but not a relatively greater increase for new variants.

The new pre-print by David Ho’s group linked at top of this thread reports what happens w new booster that was rolled out this fall (2023), which contains just XBB.1.5
Punchline is things look much better in terms of boosting neutralizing titers specifically to new variants
Punchline is things look much better in terms of boosting neutralizing titers specifically to new variants
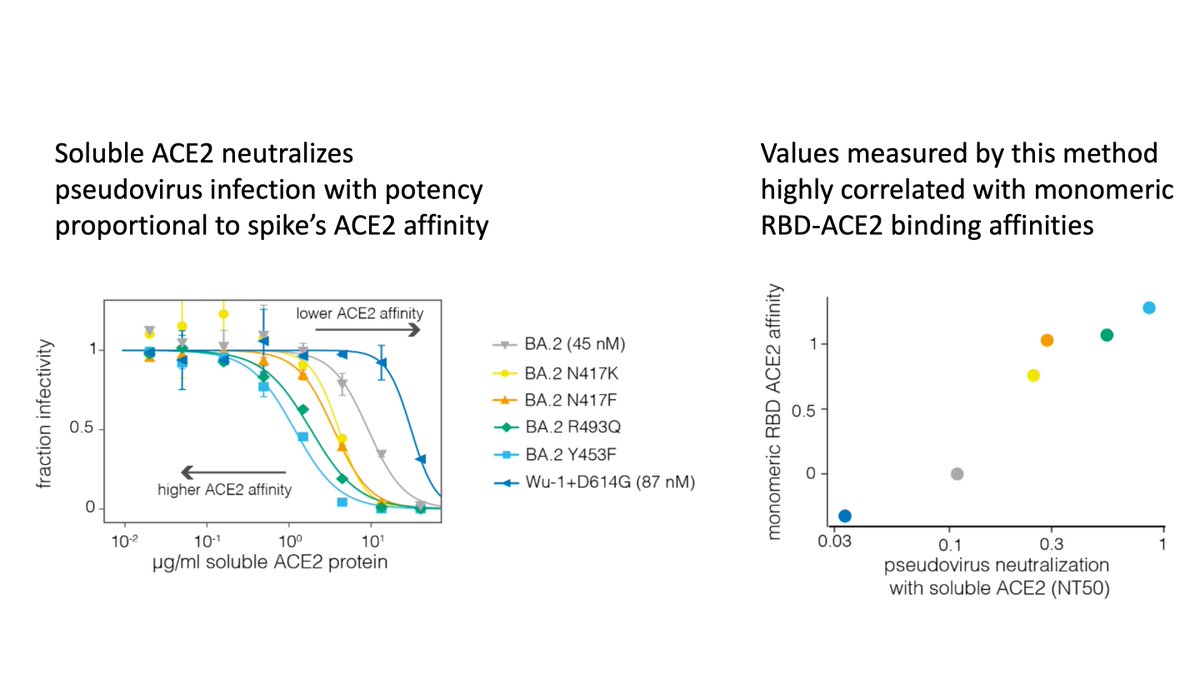
As figure below shows, XBB.1.5 booster strongly (~20-fold) increases neutralization of XBB.1.5 & XBB-descended viruses
Boost mostly specific to newer variants, as increases smaller for older variants like D614G or BA.5
Also, XBB.1.5 vaccine and XBB infection give similar boost
Boost mostly specific to newer variants, as increases smaller for older variants like D614G or BA.5
Also, XBB.1.5 vaccine and XBB infection give similar boost

Therefore, new XBB.1.5 booster is much better than prior booster at specifically boosting titers to newer variants.
So at serological level, this new booster is better overcoming “imprinting” (but more on that concept below).
This is a good thing.
So at serological level, this new booster is better overcoming “imprinting” (but more on that concept below).
This is a good thing.

Why is XBB.1.5 booster better boosting titers to new variants?
Current data insufficient to be sure. “Imprinting” used to refer to both serological effects (as above) or activation of new vs pre-existing B-cells
We don’t yet know what is happening at B-cell level w new booster
Current data insufficient to be sure. “Imprinting” used to refer to both serological effects (as above) or activation of new vs pre-existing B-cells
We don’t yet know what is happening at B-cell level w new booster
But to list some hypotheses:
Hypothesis 1⃣: Maybe problem w 2022 bivalent BA.5 booster was simply that inclusion of original strain impeded response to new strain, and main improvement of new booster is not including older strain.
Hypothesis 1⃣: Maybe problem w 2022 bivalent BA.5 booster was simply that inclusion of original strain impeded response to new strain, and main improvement of new booster is not including older strain.
Hypothesis 2⃣: Maybe greater antigenic distance of XBB.1.5 helps overcome imprinting possibly by reducing epitope masking by pre-existing serum antibodies, a la this study by @victora_lab nature.com/articles/s4158…
Hypothesis 3⃣: Maybe XBB.1.5 mostly still boosting “imprinted” B-cells at cellular level, but greater antigenic distance or accumulation of Omicron exposures is causing more expression & affinity maturation of antibodies that neutralize newer variants ()
https://twitter.com/jbloom_lab/status/1573405271090958337
But regardless of cause, observation that updated booster is now strongly boosting neutralization to new variants is good news for overall strategy of regularly updating vaccine to keep pace with SARS-CoV-2 evolution.
This fact is important, because evolution has not stopped. XBB-descended variants are still dominant most places globally and in USA, but there are indications that could potentially change fairly soon ().
https://twitter.com/siamosolocani/status/1729110596174385390
Descendants of highly divergent BA.2.86 variant like JN.1 growing rapidly (), and also now recombinants w BA.2.86 spike and other genomic regions from XBB-descended viruses. These BA.2.86-descended viruses have many spike mutations relative to XBB.1.5.
https://twitter.com/mvankerkhove/status/1727341769631965258
As shown by @yunlong_cao () and also the new David Ho pre-print, the BA.2.86 descendant JN.1 has the most neutralization escape of any variant, although it’s only modestly worse than newest XBB descended variants.
https://twitter.com/yunlong_cao/status/1714694253530804588
Fact XBB.1.5 booster still increases titers to JN.1 is good. But boost smaller than for XBB-descended variants, underscoring JN.1 is antigenically distinct
So there will certainly be more updated boosters in years to come; good to see evidence current one is working as hoped
So there will certainly be more updated boosters in years to come; good to see evidence current one is working as hoped

• • •
Missing some Tweet in this thread? You can try to
force a refresh

 Read on Twitter
Read on Twitter