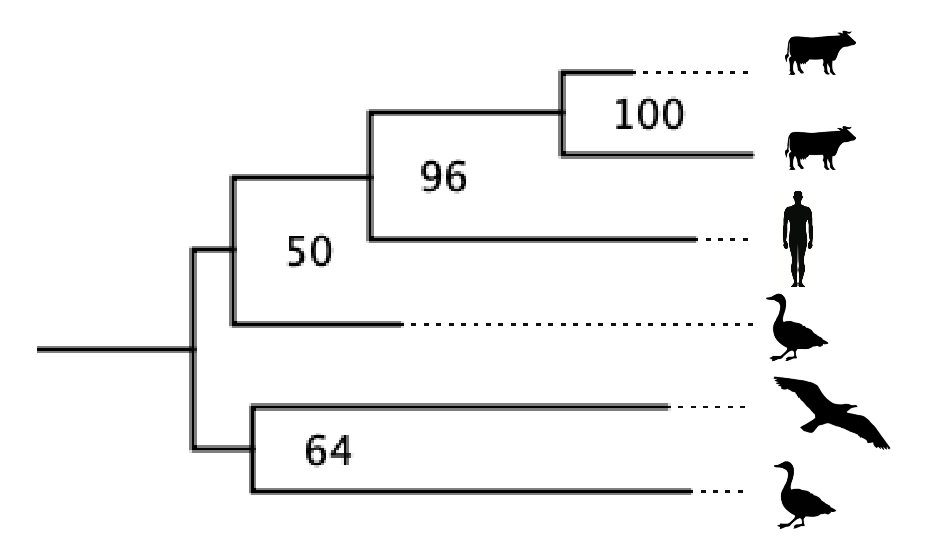
Important update on metadata of H5N1 in cattle (and back to birds):
Thanks to the extraordinary detective skills of @flodebarre, we are pleased to be able to share this table containing locations and dates for several H5N1 cases in cattle and birds:
github.com/andersen-lab/a…
Thanks to the extraordinary detective skills of @flodebarre, we are pleased to be able to share this table containing locations and dates for several H5N1 cases in cattle and birds:
github.com/andersen-lab/a…
We have pseudomized specific location data relating to individual farms/herd/operations, and are only sharing location to state.
We are now incorporating this important metadata into our phylogeographic analyses, which will allow us to do things like use "local molecular clocks" of the sort that Andrew Rambaut and I previously used to resolve the deep history of influenza A virus:
nature.com/articles/natur…
nature.com/articles/natur…
Sidebar: this paper also discusses the "Great Epizootic of 1872".
Haven't heard of it? It's a fascinating story.
Haven't heard of it? It's a fascinating story.
We will also be attempting to pin down the most likely location of origin of this outbreak,. And we are integrating sequence, timing, and geographical information to investigate how these viruses have moved and are moving within and between states.
Please note that, in addition to the metadata, the consensus sequences from our genome assemblies of the 239 cases times 8 genome segments are publicly downloadable from our github.
As our our sequence alignments.
github.com/andersen-lab/a…
As our our sequence alignments.
github.com/andersen-lab/a…
Thanks to Andy Bowman and @Jimpigvet for sharing consensus genomes from H5N1 cases they sampled in Ohio.
And to all the people collaborating on our efforts, including @xrayfoo @flodebarre Kristian Andersen @LouiseHMoncla @swientist @meera_chand @MOUGK @EvolveDotZoo @stgoldst @stuartjdneil @PeacockFlu Andrew Rambaut @angie_rasmussen @suchard_group @LemeyLab @jepekar @josh__levy
Joel Wertheim @LrnM9 @evogytis Daniel Goldhill Chris Ruis
As well as @USDA_APHIS for making their SRA data available, and all the veterinarians, farmers, farmworkers, and others doing the boots-on-the ground work here.
As well as @USDA_APHIS for making their SRA data available, and all the veterinarians, farmers, farmworkers, and others doing the boots-on-the ground work here.
@jimpigvet And thanks to Rich Webby too!
• • •
Missing some Tweet in this thread? You can try to
force a refresh