Finally out! Miriam's (@miriamppol) 1st, 1st author article!
It came out for the international day of women in science! Miriam is one of my first PhD students co-supervised with @Alfons_Valencia!
1st last author paper as a group leader! 🥹
Thread🧵 doi.org/10.64898/2026.…
It came out for the international day of women in science! Miriam is one of my first PhD students co-supervised with @Alfons_Valencia!
1st last author paper as a group leader! 🥹
Thread🧵 doi.org/10.64898/2026.…

We had a fundamental question that Burkhard Rost has addressed decades ago!
How large is the sequence attractor of a given protein fold?
Which positions can be varied in sequence so that the fold does not care and which ones are not changeable?
(+)
How large is the sequence attractor of a given protein fold?
Which positions can be varied in sequence so that the fold does not care and which ones are not changeable?
(+)
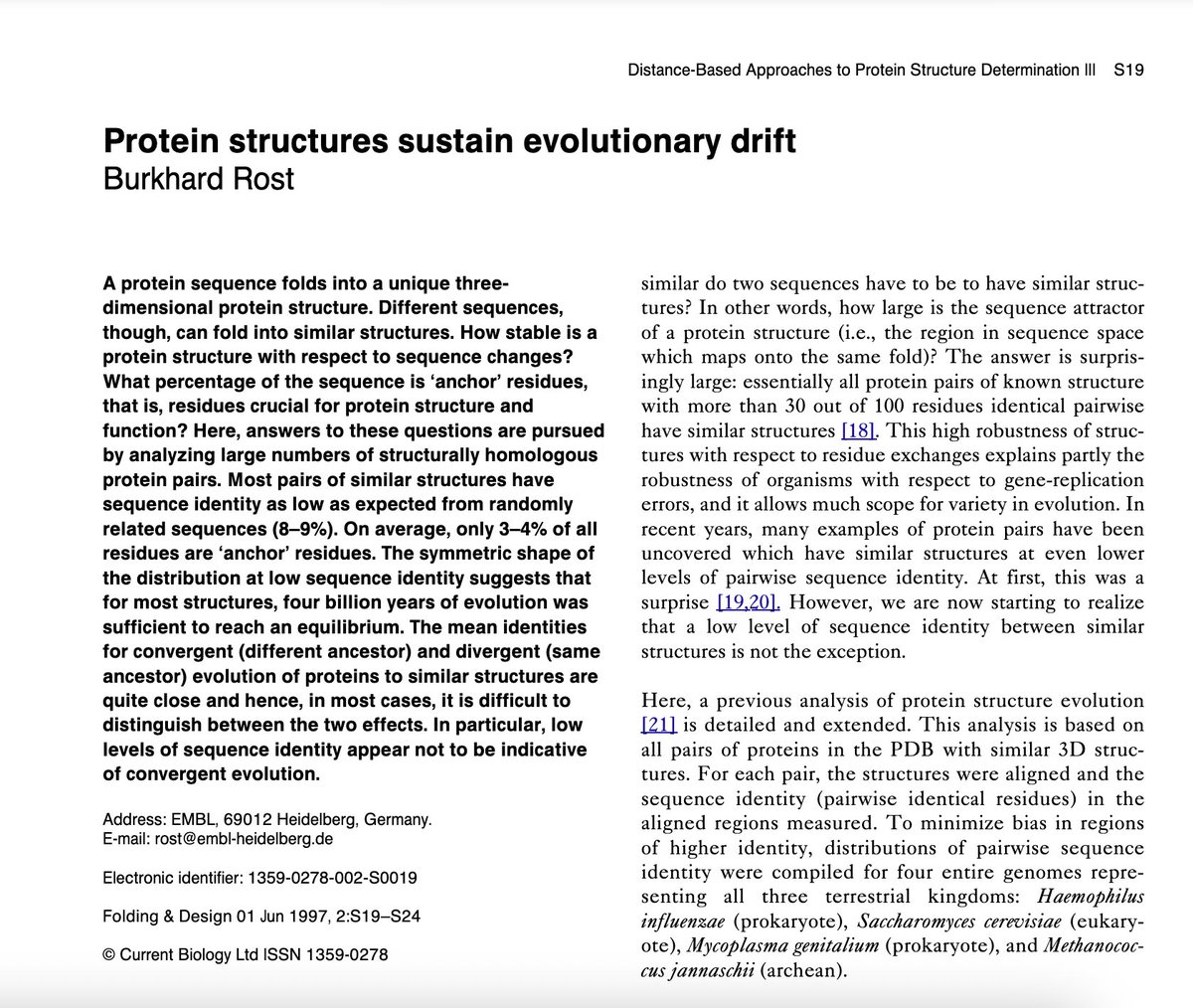
Using reverse-folding algorithms (ProteinMPNN, Caliby) + structure prediction + local frustration analysis,
we redesigned sequences for fixed backbones.
We used FrustraEvo to analyse:
Which positions are free to vary and which are energetically constrained?
(+)
we redesigned sequences for fixed backbones.
We used FrustraEvo to analyse:
Which positions are free to vary and which are energetically constrained?
(+)
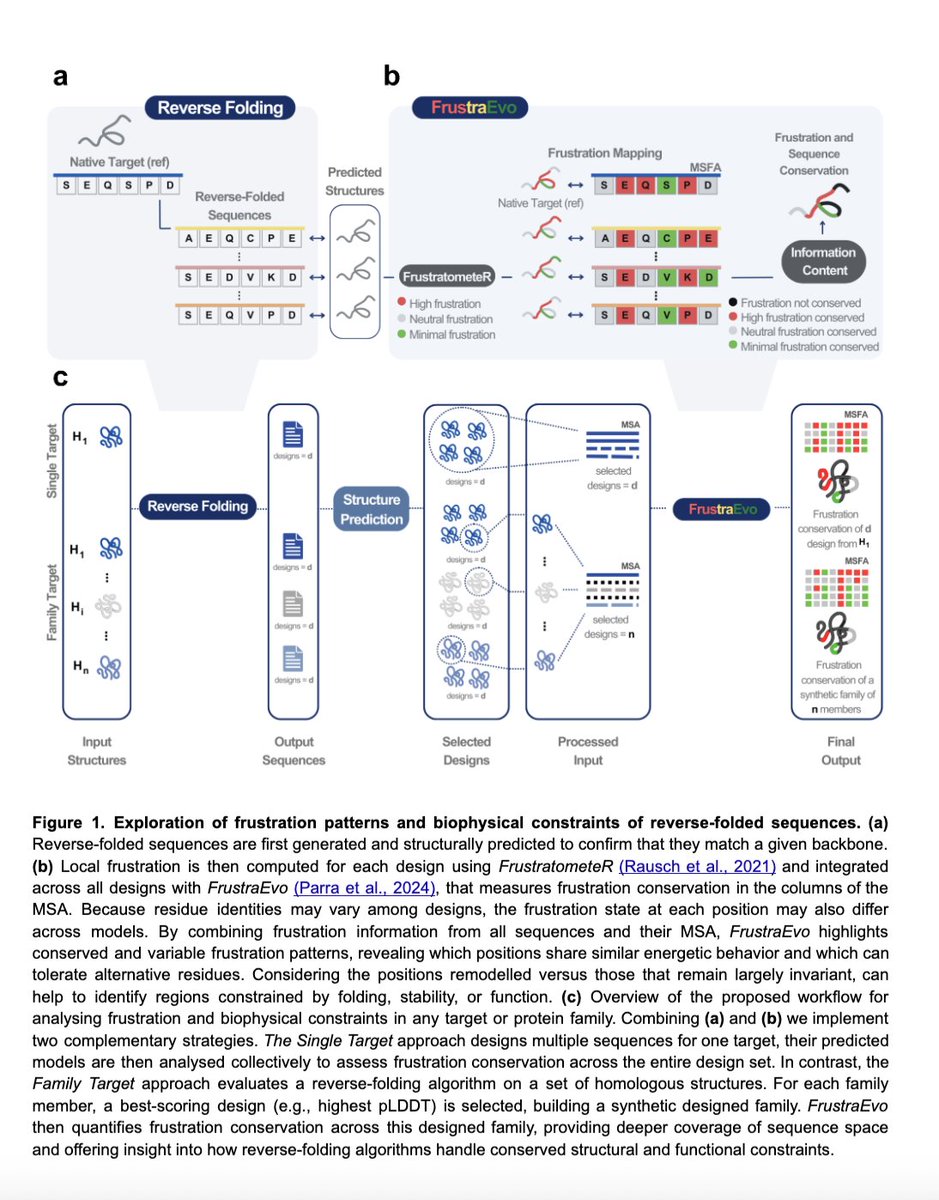
We analyzed this in the context of alpha-globins where protein-protein interaction sites, known to be highly frustrated. Sequences are remodelled to largely decrease frustration!
That makes sense as ProteinMPNN maximises stability which conflicts with function.
(+)
That makes sense as ProteinMPNN maximises stability which conflicts with function.
(+)
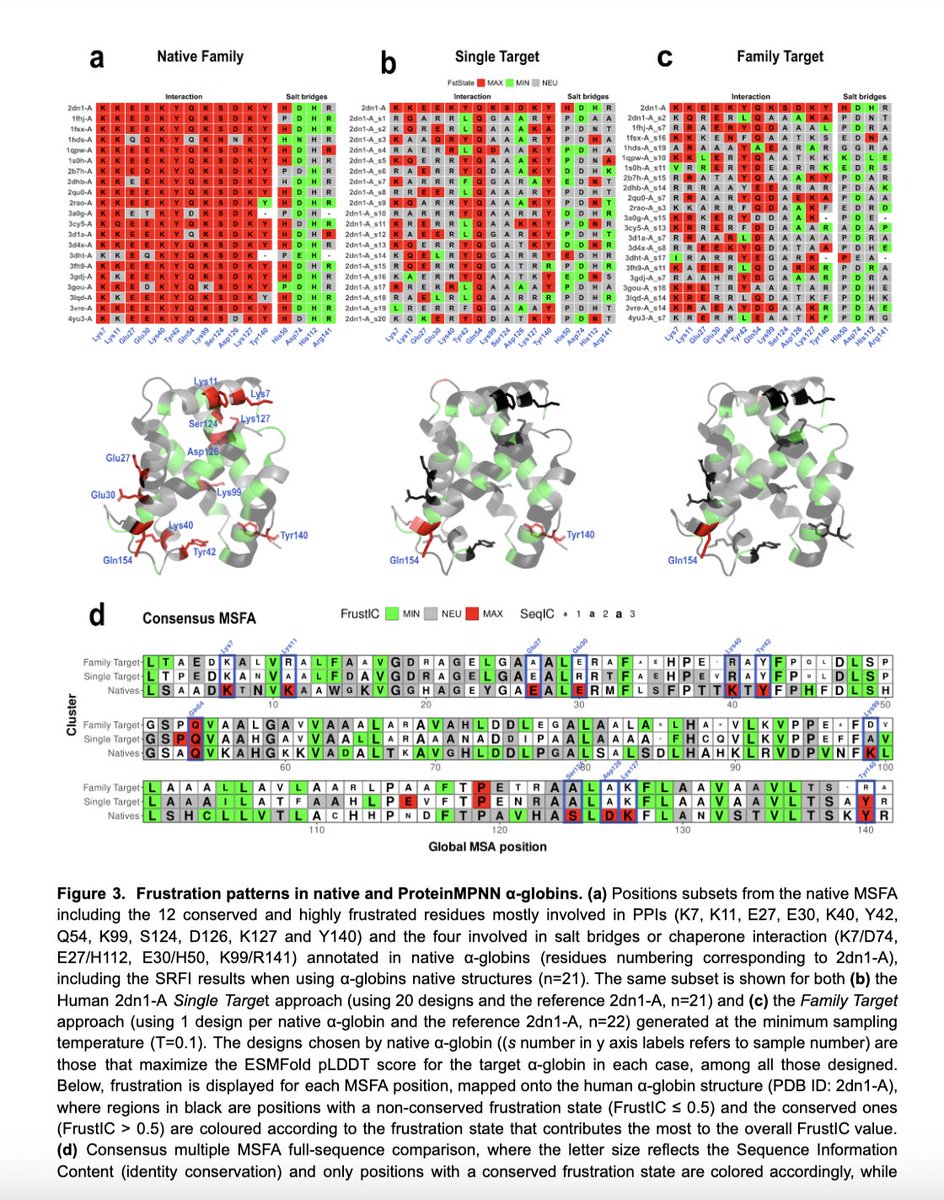
Our next family to analyse was Beta-Lactamases, whose catalytic sites are also known to be highly frustrated.
To our surprise, reverse folded sequences were not remodelled. The native, highly frustrated identities are always recovered in the design.. This made no sense...!
(+)
To our surprise, reverse folded sequences were not remodelled. The native, highly frustrated identities are always recovered in the design.. This made no sense...!
(+)
Why are reverse folded sequences maintaining energetic conflicts if they are maximising stability and the sequence-to-structure fit? Is there a bias in the methods? Has ProteinMPNN memorized catalytic sites and just imprints its identities when seeing something alike? (+)
We mutated in silico all catalytics to to Valines to explicitly minimise frustration. We also used experimentally mutated structures. Reverse folded sequences using both methods still recovered the native identities back.. Is there a memory in ProteinMPNN? 
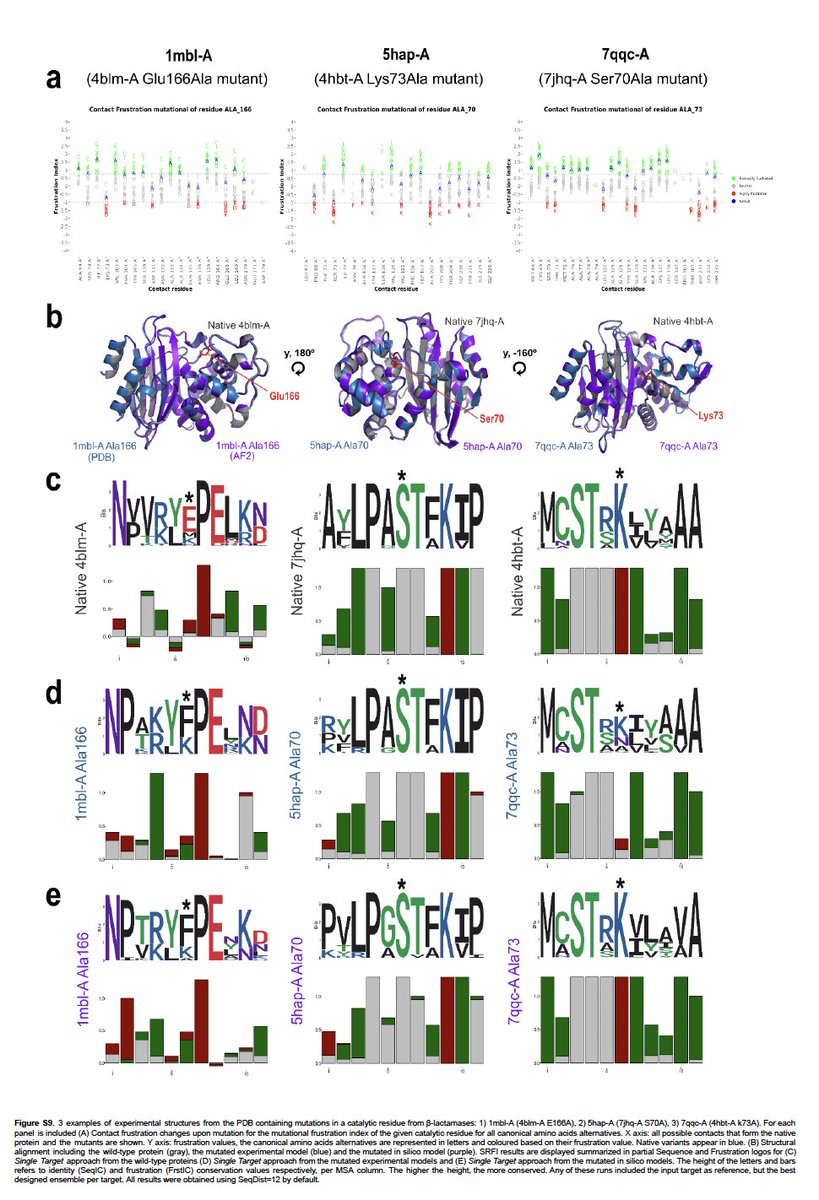
We pushed ProteinMPNN to its limits. We used max temperature to maximise seq varability. We also retrained it by deleting from the training set all annotated and predicted enzymes!
Some signal was gone but some was not! It this still evolutionary information leakeage?
(+)
Some signal was gone but some was not! It this still evolutionary information leakeage?
(+)
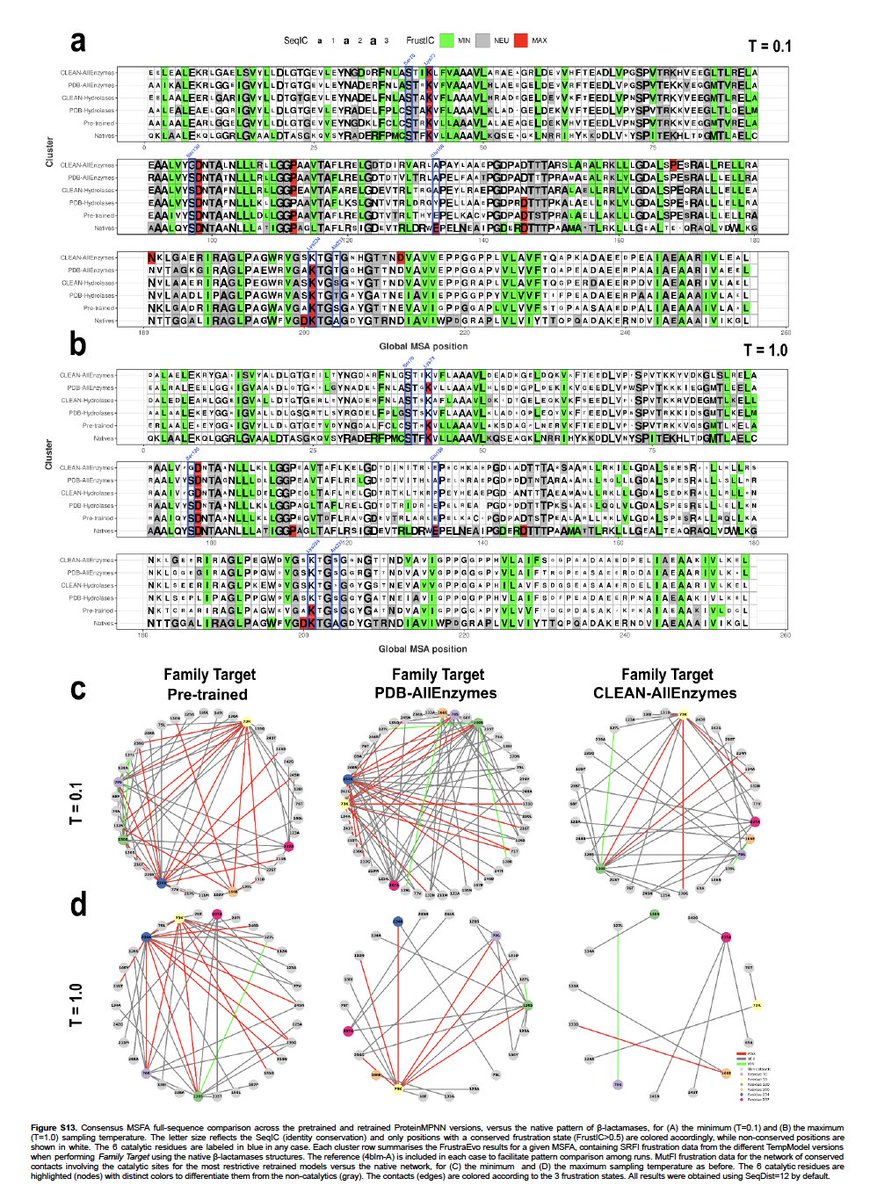
We then decided to use De Novo designed folds as they are designed to maximise foldability, stability and have not (known) function.. Top7 is very intersting.
Both the original design and the ProteinMPNN designs (Baker vs Baker metaverse) contain highly frustrated residues (+)
Both the original design and the ProteinMPNN designs (Baker vs Baker metaverse) contain highly frustrated residues (+)
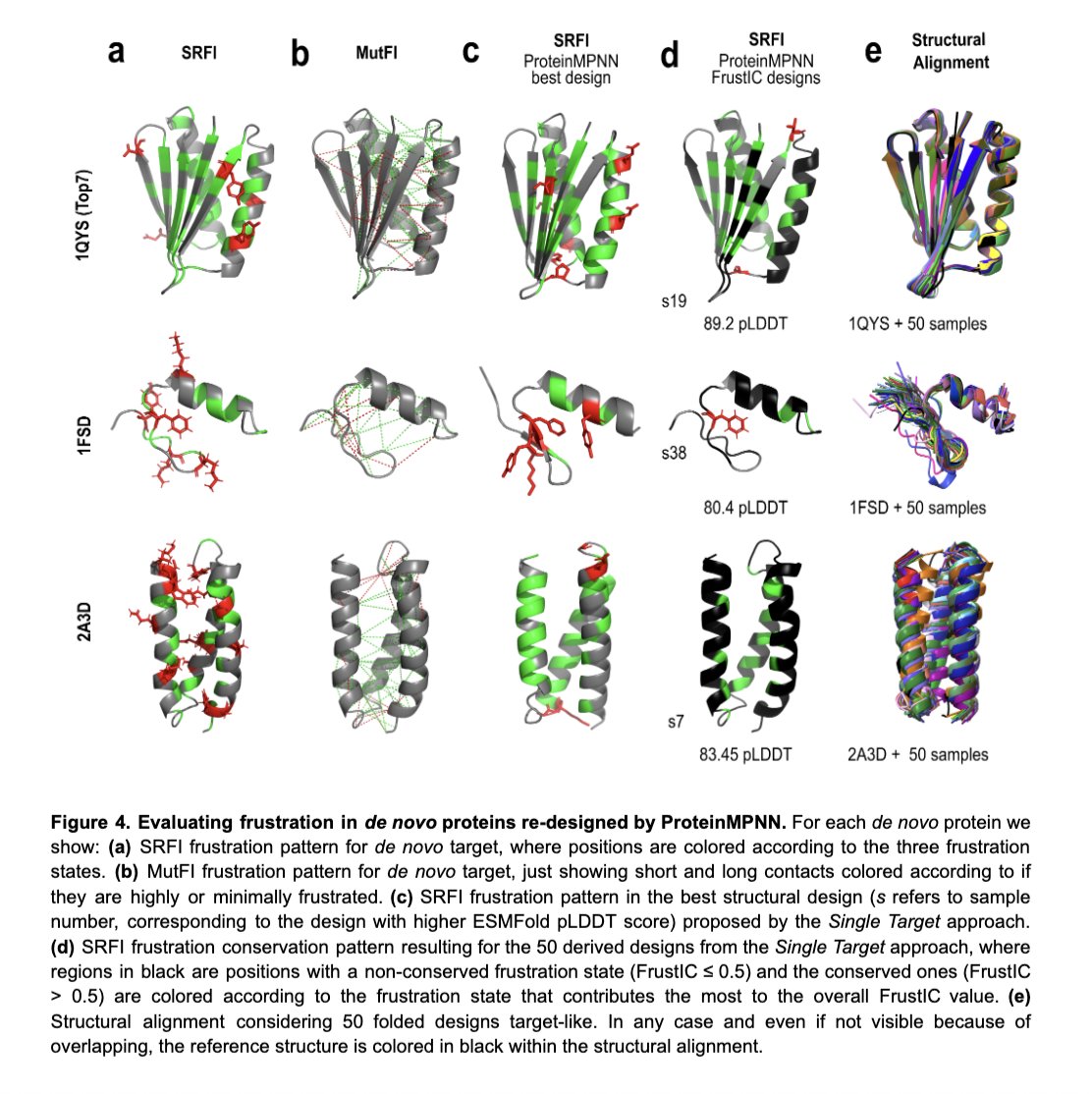
We conclude that some frustration that cannot be erased from the sequences even when reverse folding to maximise sequence-structure-fit behaves as a spandrel. This frustration is not the consequence of adaptive evolution but a consequence of the fold architecture.
(+)
(+)
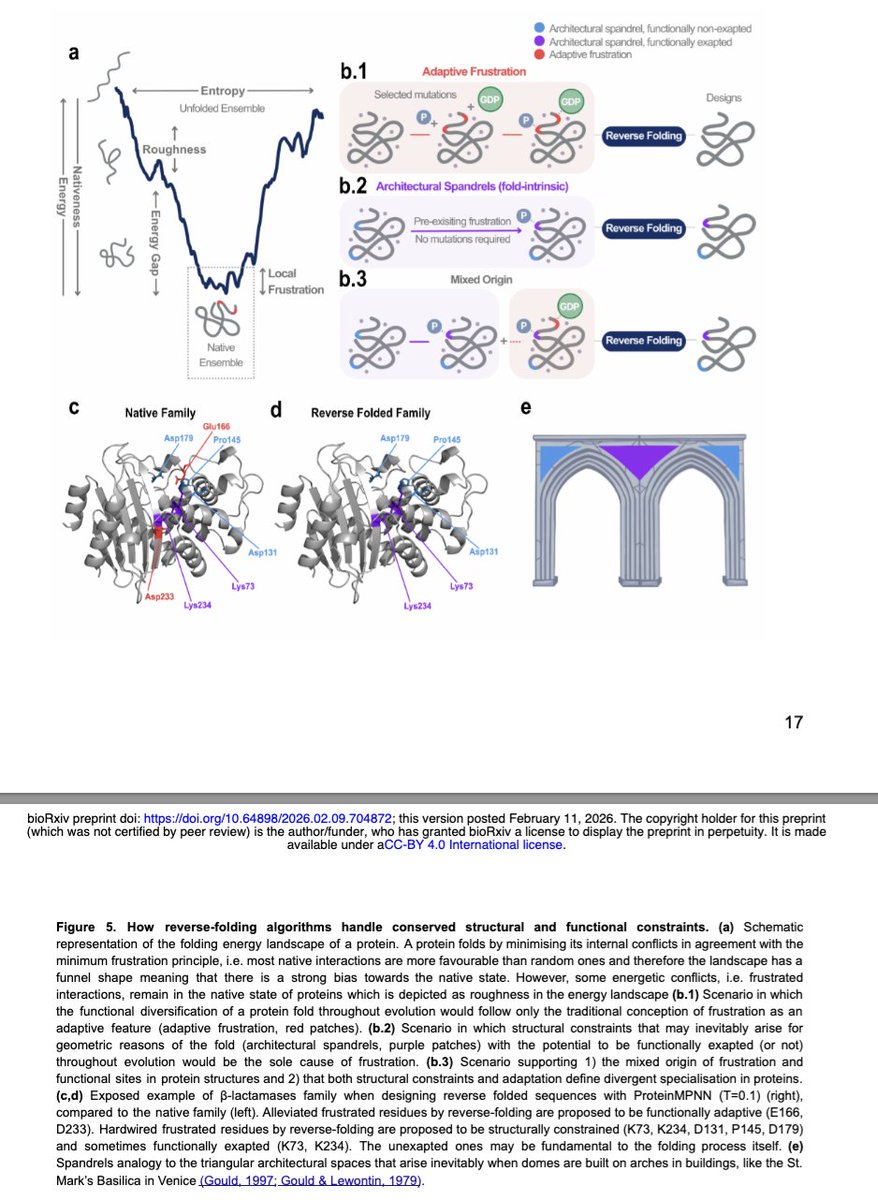
On his postume article, Dan Tawfik & colleagues propose that ancestral enzymatic function could be seeded by unstable hotposts in proteins that could bind small molecules such as phosphate with low affinities by separating them from the environment... (+) 
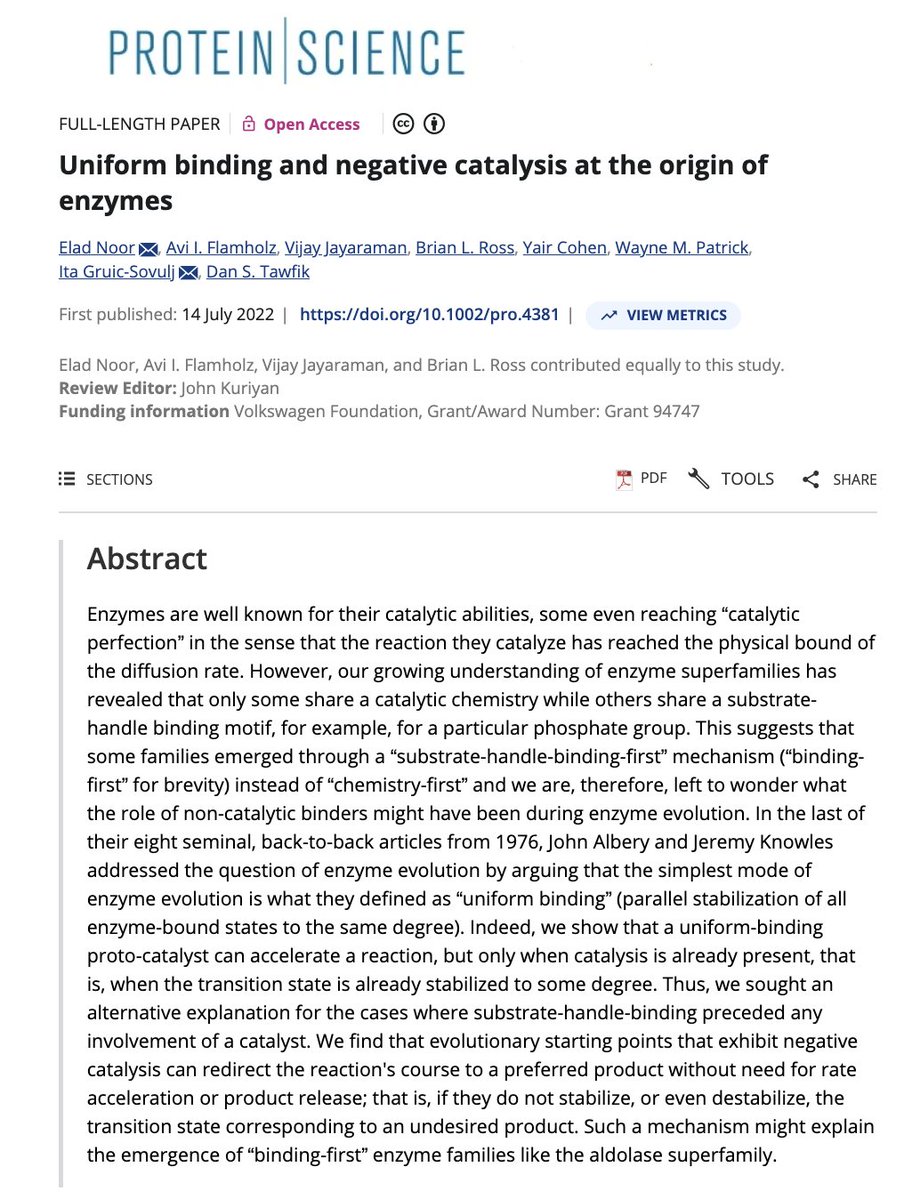
Such sites could have later evolded to become more complex and give rise to modern catalytic sites. We know that frustration can represent those ancestral hotpots. A spandrel that has no reason other than to facilitate a complex structure can be that ancestral hotspots (+)
This idea is also compatible with the theory of platonic folds. Sequences don't really code structures but fall into attractors defined by folds. Folds are the consequence of physical and biochemical rules given the available amino acids and how they interact with solvent (+) 

Folds represent basins in sequence space & sequences diffuse such space until the fall into one of this folds. We know that sequences are not evenly distributed across structure space. Something shown by Christine Orengo years ago. Are superfolds, super platonic attractors? (+) 
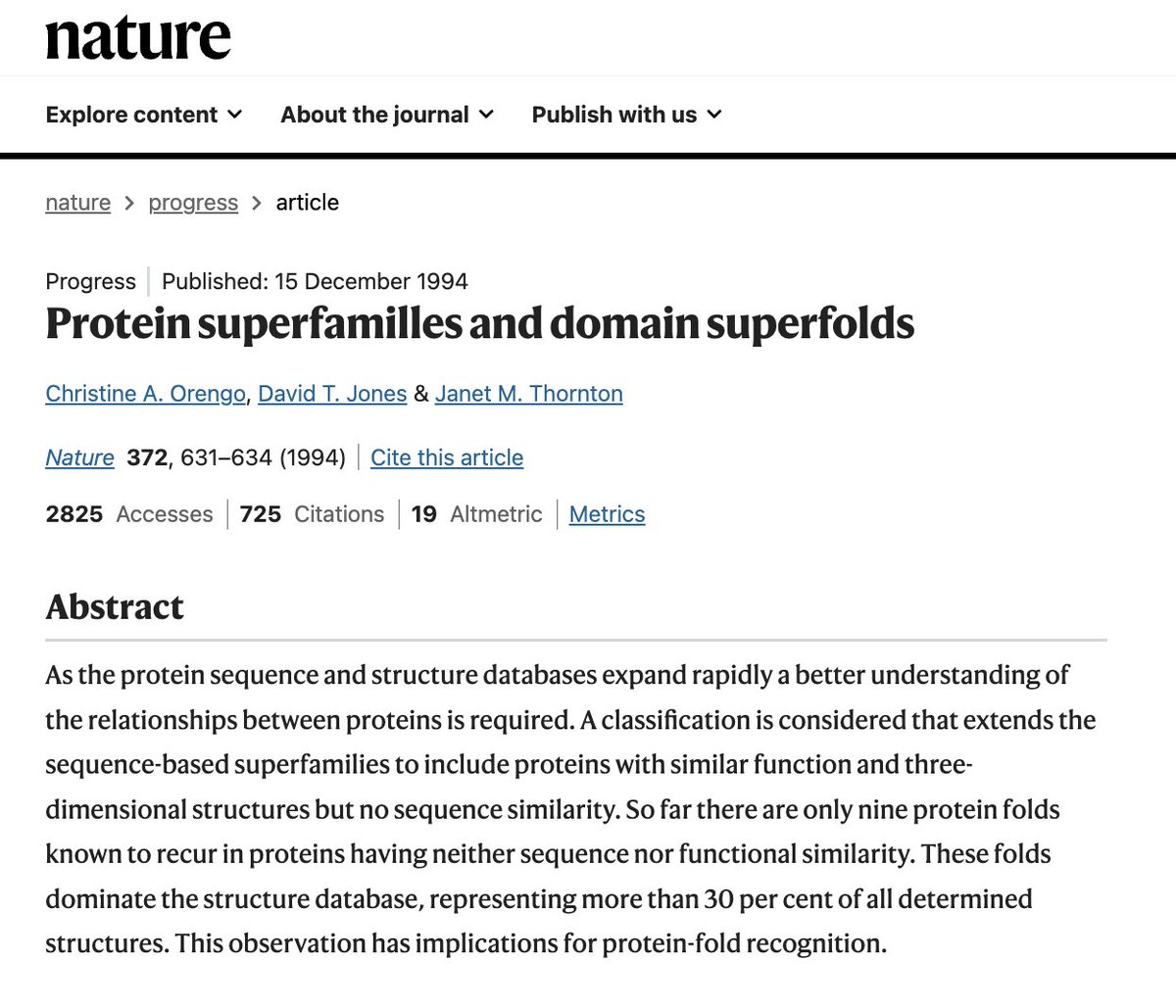
If you want a complex architechture it makes sense that you cannot have a completely frustration free structure. You need "hinge" residues so you can adopt such folds. Maybe these are our frustration spandrels that can be later on exapted for function.. as Gould proposed (+) 

Our study implied 1000s of predicitons & calculations but still it only represents few folds as case studies. Is this something more general? We will study all known enzyme families to complete this idea but we want to present this initial work as potential evidence (+)
This has been a tremendous work by Miriam and collabs. It is my first paper as a group leader so it scares me a bit.. but it was a great adventure! Let's see what reviewers say! Comments Welcome! (+)
We have recently lost 2 great scientists in our field. Amos Bairoch & Peer Bork who not only have inspired my work since my early years but also built theory, tools & databases without which this work would not have been possible. This work is dedicated to their memory. RIP❤️.
@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh










