Avid genome spelunker | @HarvardMed Ph.D. candidate | Home = @talkowskilab at @CGM_MGH & @BroadInstitute | @NSF fellow | @dartmouth alumnus | Opinions my own
How to get URL link on X (Twitter) App

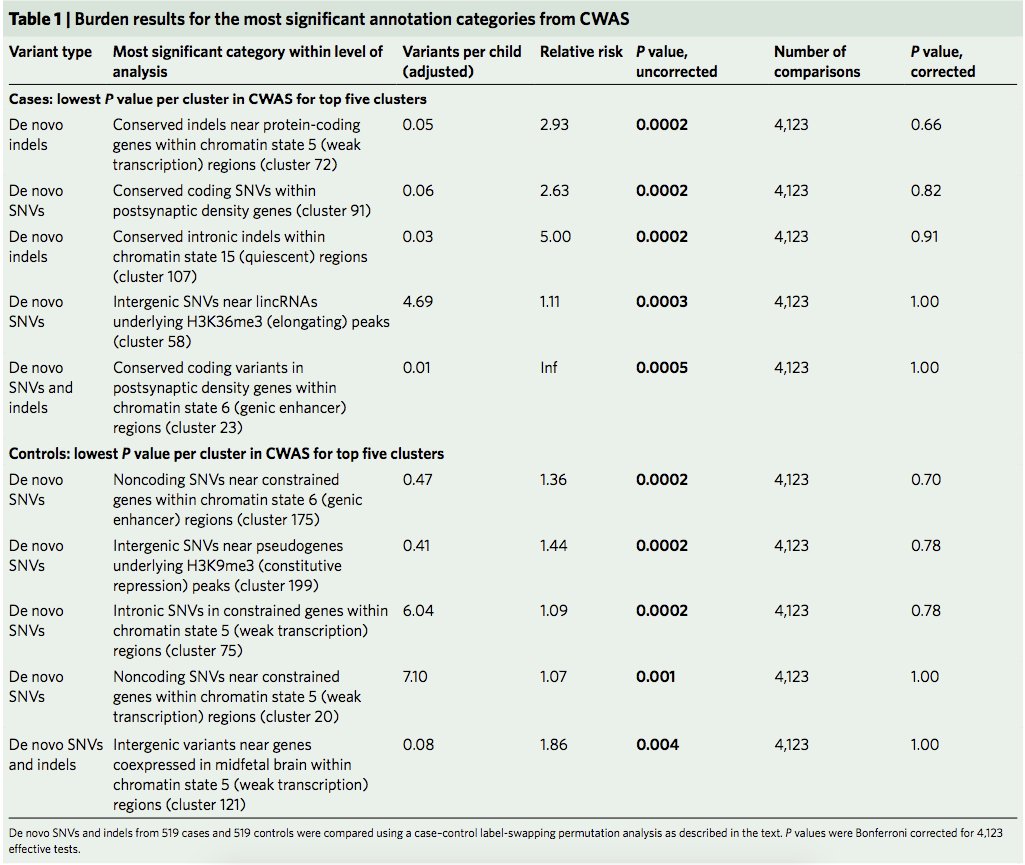
 1. We compared effects of rare CNVs (rCNVs) across 54 disorders
1. We compared effects of rare CNVs (rCNVs) across 54 disorders