"Landscape and Selection of Vaccine Epitopes in SARS-CoV-2"
(biorxiv.org/content/10.110…)
Collaboration with @ChristofCSmith1 @bgvincentlab @JuliaKodysh @timodonnell @glycam @Olivercgrant @PEPperCHIP @erikgarrison & others.
Thread:
(1/)
Front-runners: inactivated virus (SinoVac), mRNA (BioNTech, Moderna), electroporated DNA (Inovio), adenovirus (CanSino, Oxford), rVSV (IAVI), rS (Novavax) &c
Missing from the list: peptides
2/
Peptide vaccines aren't often used for infectious disease because they can only target linear B-cell epitopes.
3/
nature.com/articles/s4157…
4/
5/
6/
Which brings me to the subject of our preprint...
7/
8/
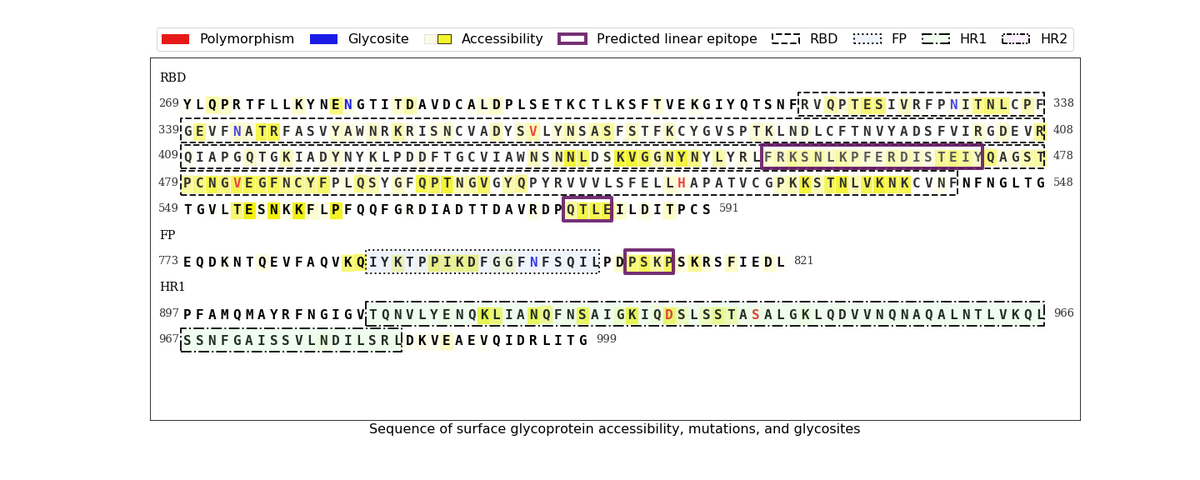
T-cell: MHC binding, immunogenicity, exclude polymorphic sites, abundant proteins (M, N, S).
B-cell: use linear B-cell epitopes from patient serum, filter polymorphic sites, glycosites, inaccessible regions.
Vaccine: pick long peptides with multiple epitopes
9/
To further refine the set we made a model of p(immunogenic|MHC ligand) from IEDB's viral pMHC tetramer results
11/
12/
(e.g. smaller AAs = less likely)
13/
One unpublished peptide array study from @PEPperCHIP (which helped us get started), 3 published peptide mapping studies, and one PhIPseq paper: medrxiv.org/content/10.110…
16/
S456-473 (RBM loop)
S580-583 (near RBD)
S809-812 (near FP)
The last two were the targets of neutralizing antibodies in biorxiv.org/content/10.110…
& the last B-cell epitope region occurred in all 5 sources!
18/

21/
I'm really excited for the next round of experiments...
22/
Tune in for the next paper...
23.
We started with measured B-cell epitopes (albeit of unknown function) but the T-cell epitopes were purely computational predictions. Since we did these analyses, 2 preprints came out which identify T-cell epitopes in recovered SARS-CoV-2 patients...
24/
25/
(medrxiv.org/content/10.110…)
also found 2 recurrent epitopes (S269-277, S1000-1008). These are also contained in our vaccine peptide set
(5B, peptides 11 & 15), So far so good!
26.









