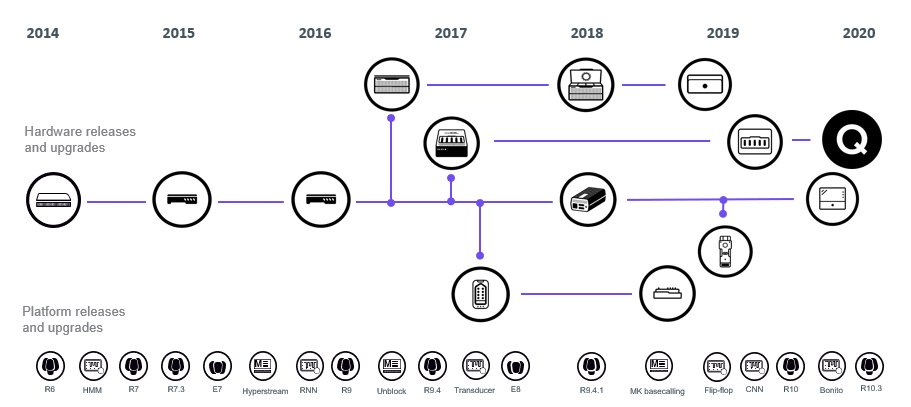
#nanoporeconf
#nanoporeconf
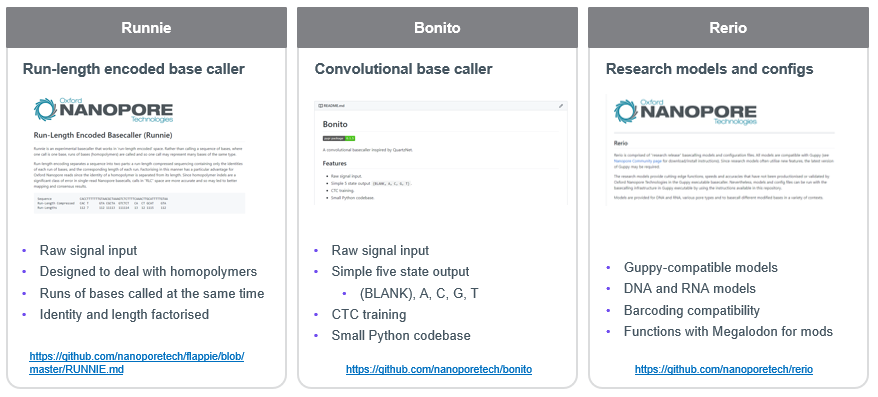
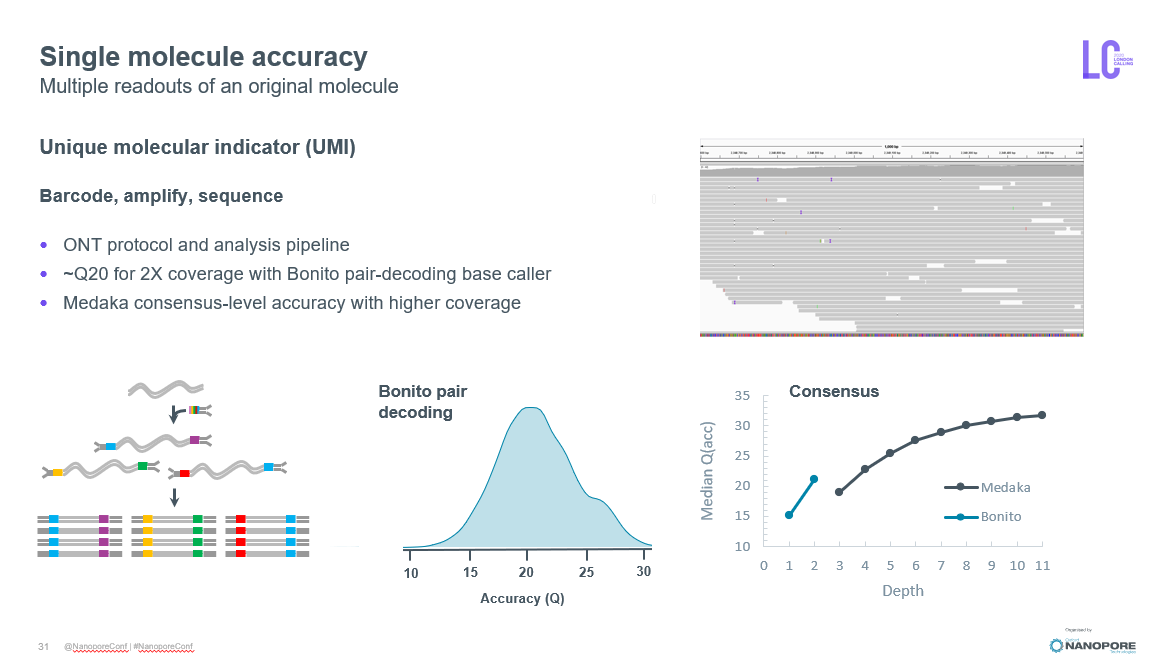
for microbial community #nanoporeconf nanoporetech.com/accuracy
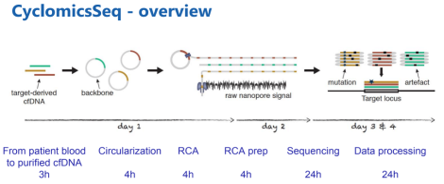
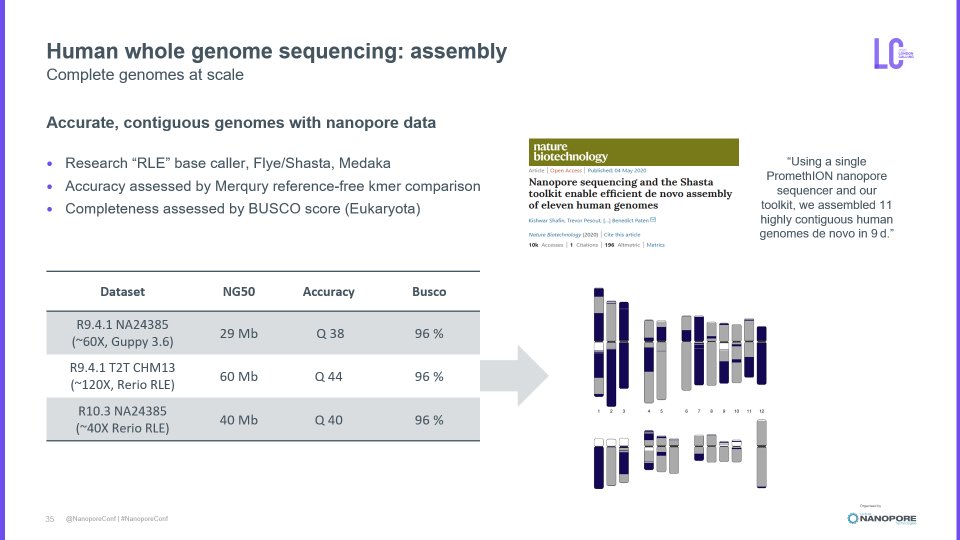
#nanoporeconf
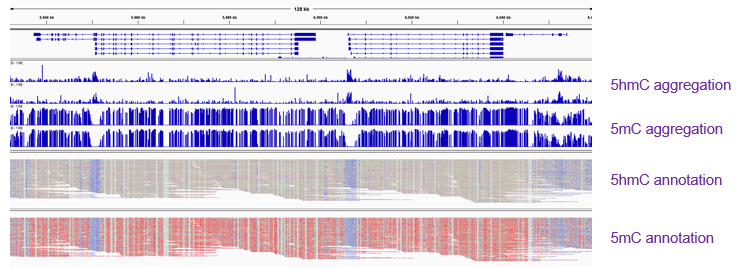
#nanoporeconf - you can hear more tomorrow from @mattloose too
youtube.com/watch?v=fwHreH…
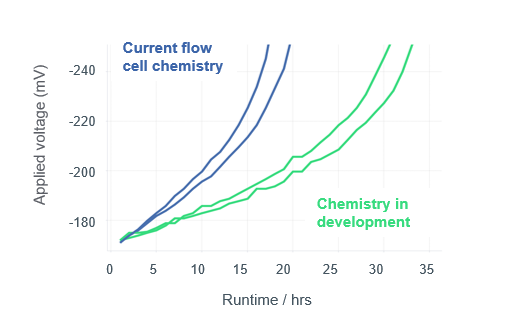
#nanoporeconf
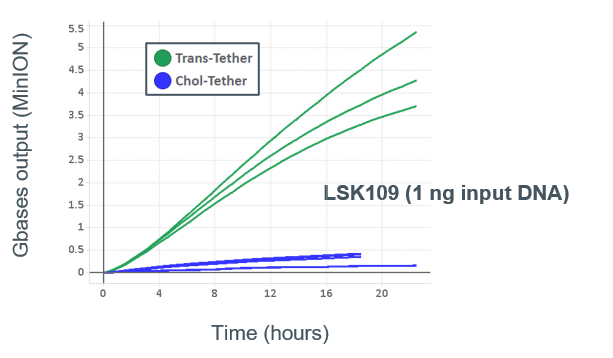
nanoporetech.com/products/plong…

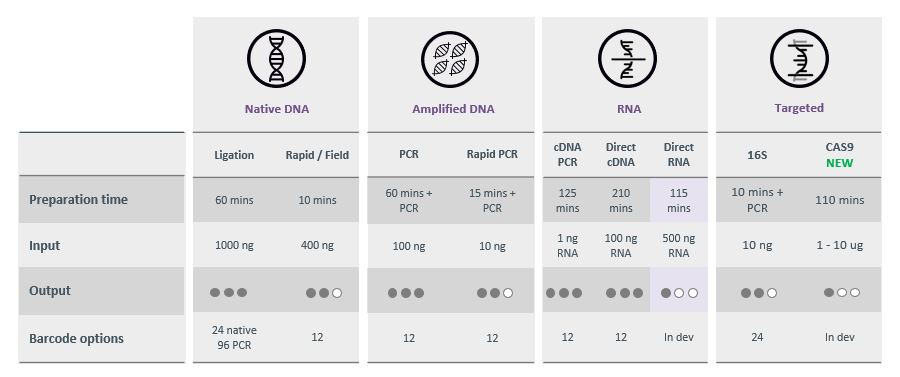
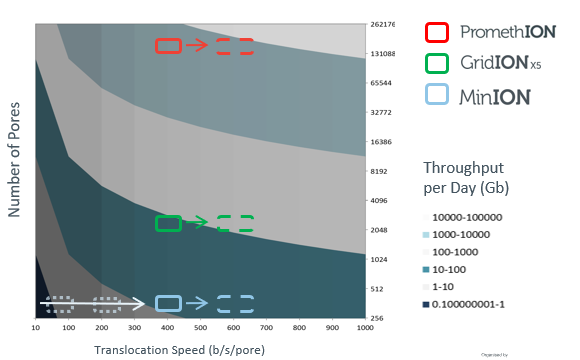
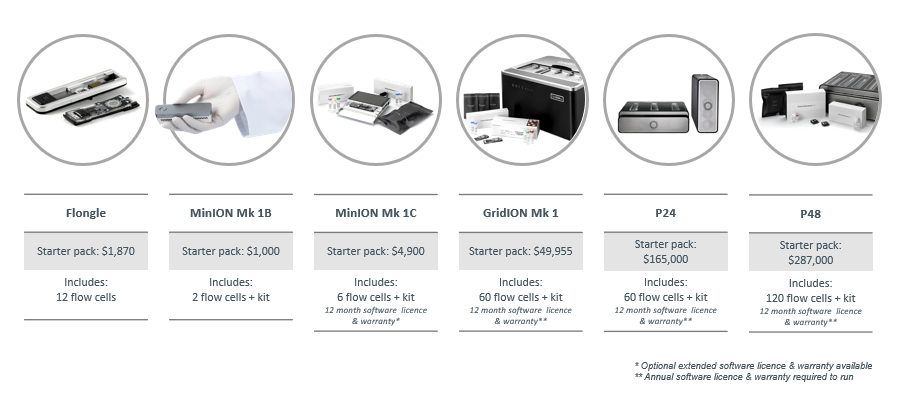
nanoporetech.com/products/compa…