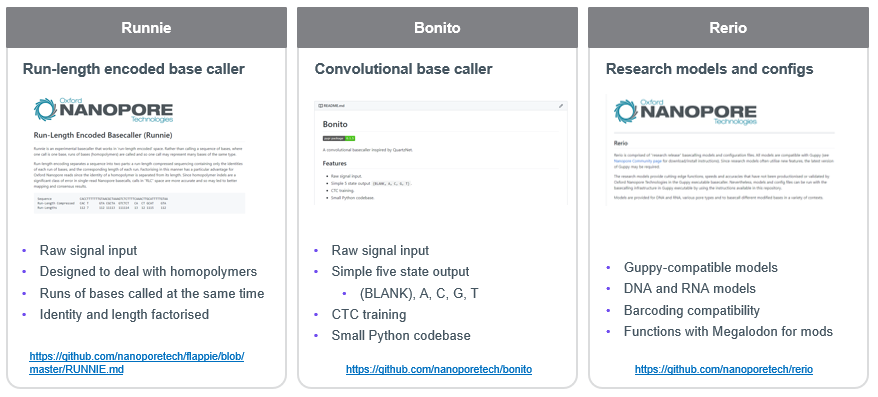
🌍Direct DNA/RNA analysis for anyone, anywhere
🧬Short to ultra-long reads in real-time for rapid insights
🔬Products for research use only
How to get URL link on X (Twitter) App


 Please note, we invite you to read the disclaimer in the slides
Please note, we invite you to read the disclaimer in the slides

 GS: Lord Kelvin believed in meaningful measurement to see the whole picture. With nanopore sequencing you gain more comprehensive insights than ever before. #nanoporeconf
GS: Lord Kelvin believed in meaningful measurement to see the whole picture. With nanopore sequencing you gain more comprehensive insights than ever before. #nanoporeconf 



 @The_Taybor CB: our goal is to enable the analysis of anything, by anyone, anywhere #nanoporeconf
@The_Taybor CB: our goal is to enable the analysis of anything, by anyone, anywhere #nanoporeconf

 Whilst the amplicon dropout does not affect identification of the Omicron variant, the Midnight kit will be revised to include an additional primer to improve amplification efficiency of Omicron Amplicon 28. We expect this to be available in 2-4 weeks 2/4
Whilst the amplicon dropout does not affect identification of the Omicron variant, the Midnight kit will be revised to include an additional primer to improve amplification efficiency of Omicron Amplicon 28. We expect this to be available in 2-4 weeks 2/4