
Almost three years of work to deliver Baysor: a tool for Bayesian segmentation of #spatial data (FISH and in-situ sequencing)! Besides the cell segmentation, we also provide a framework for segmentation-free analysis and discuss the field in general.
biorxiv.org/content/10.110…
(1/7)
biorxiv.org/content/10.110…
(1/7)
Neighbourhood Composition Vectors provide an easy way to run your scRNA-seq pipelines on spatial data without running cell segmentation (b,c) and also allow beautiful visualization of the tissue composition (a)
(2/7)
(2/7)

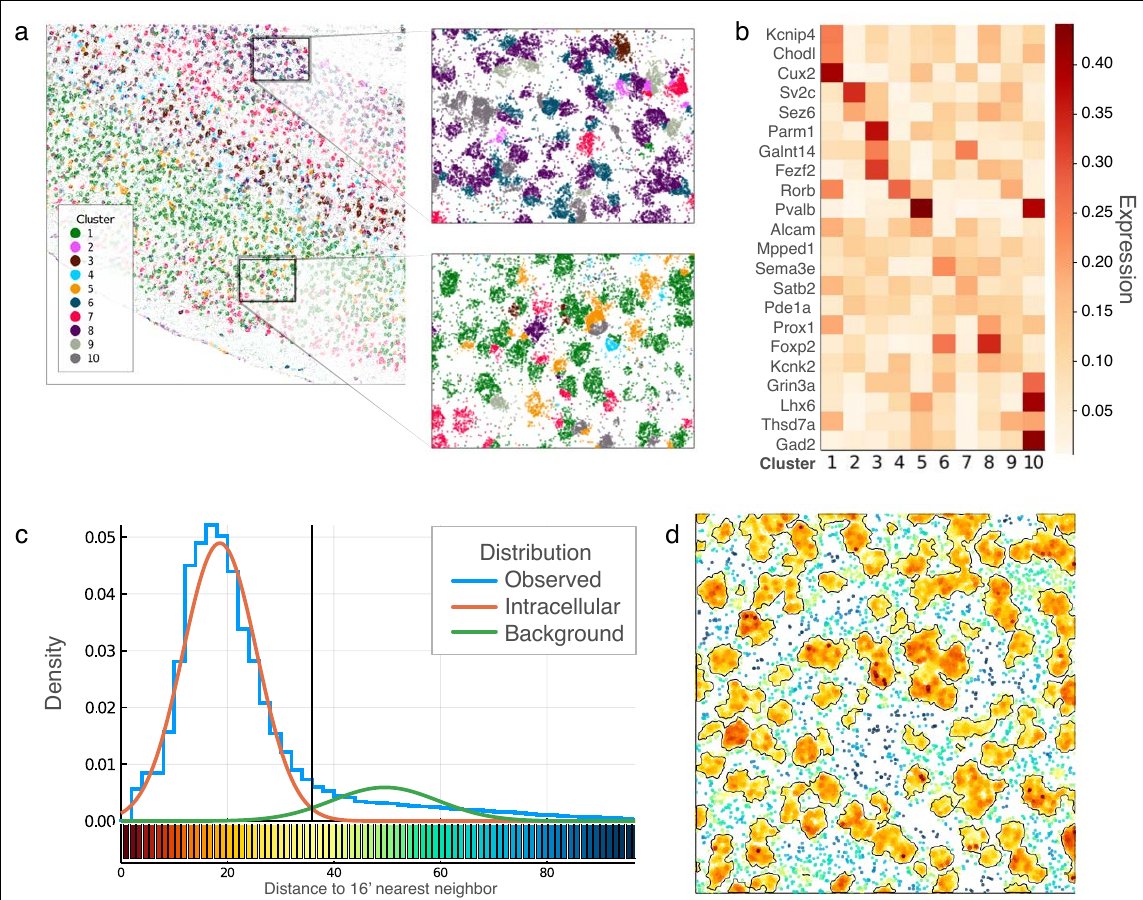
We also describe a Markov Random Field-based framework for labelling molecules. Here we apply it for ultra-fast inference of cell types (a-b) and background filtration (c-d).
(3/7)
(3/7)
Baysor cell segmentation works on most of the published protocols, including MERFISH (a), @AllenInstitute smFISH (b), ISS (c), STARmap (d) and osm-FISH (e)
(4/7)
(4/7)

With the use of #JuliaLang, full dataset segmentation can be run on laptop, working only 12 minutes single-thread on Allen smFISH (1M molecules, 22 genes) and 51 minutes on MERFISH (3.7M molecules, 140 genes).
(5/7)
(5/7)
Finally, we discuss limitations of the approach: ultra-sparse data (f), 3D structure (c), or working with super-high resolution data, such as seq-FISH+ (a). Still, most of these problems can be solved by providing a staining-based segmentation as a prior to Baysor (g-h).
(6/7)
(6/7)

We're highly grateful to @SimoneCodeluppi and @LarsBorm from @slinnarsson lab, Xiaoyan Qian and @M_Hilscher from @MatsNilssonLab, Jeff Moffit, and especially Brian Long and Bosiljka Tasic from @AllenInstitute for helping us with data.
(7/7)
(7/7)
• • •
Missing some Tweet in this thread? You can try to
force a refresh


