Any info out on the 'new strain' in phylogenetic / AA mutation terms?
Wonder if there's any chance increased circulation could be driven by false-negatives in testing. (happens in malaria with hrp2 deletion) 

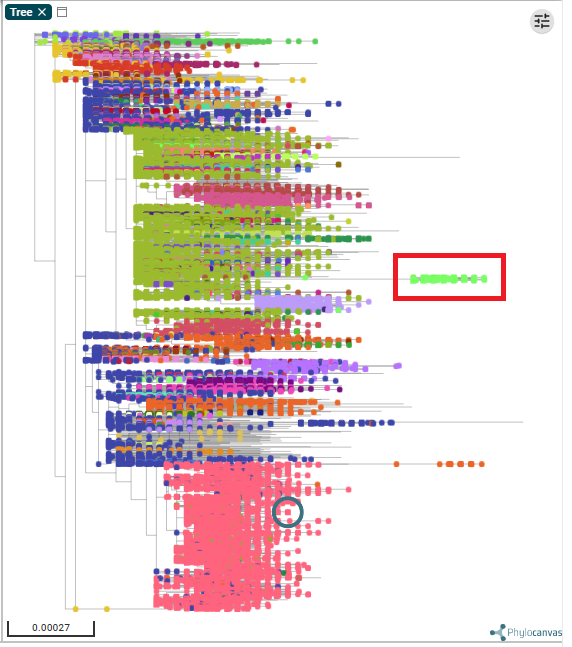
So if I understand correctly it's this clade on microreact beta.microreact.org/project/4ebHf8… 
N501Y also appeared during adaptation to mice science.sciencemag.org/content/369/65…
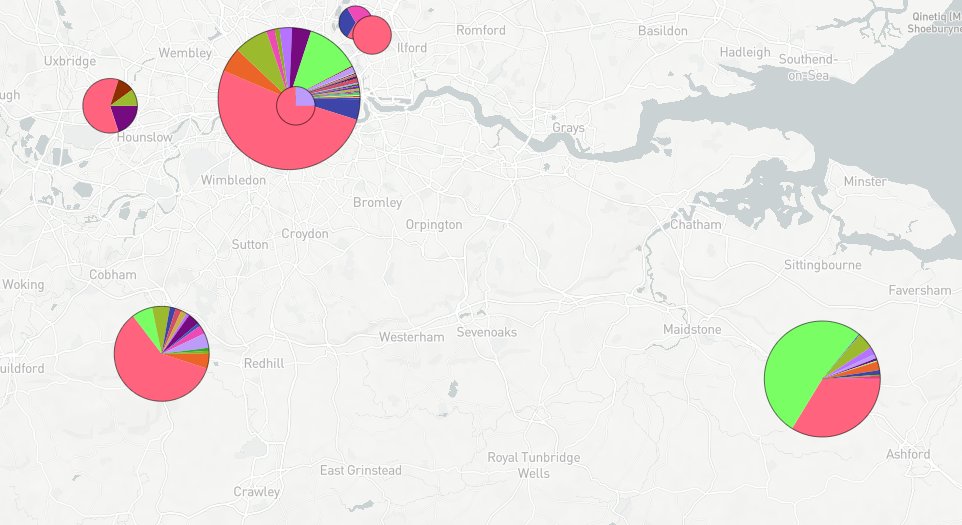
Filtering to only show samples after 15 Nov: Lime green strain making up majority of Kent cases and substantial proportion in London (and rare outside SE) 
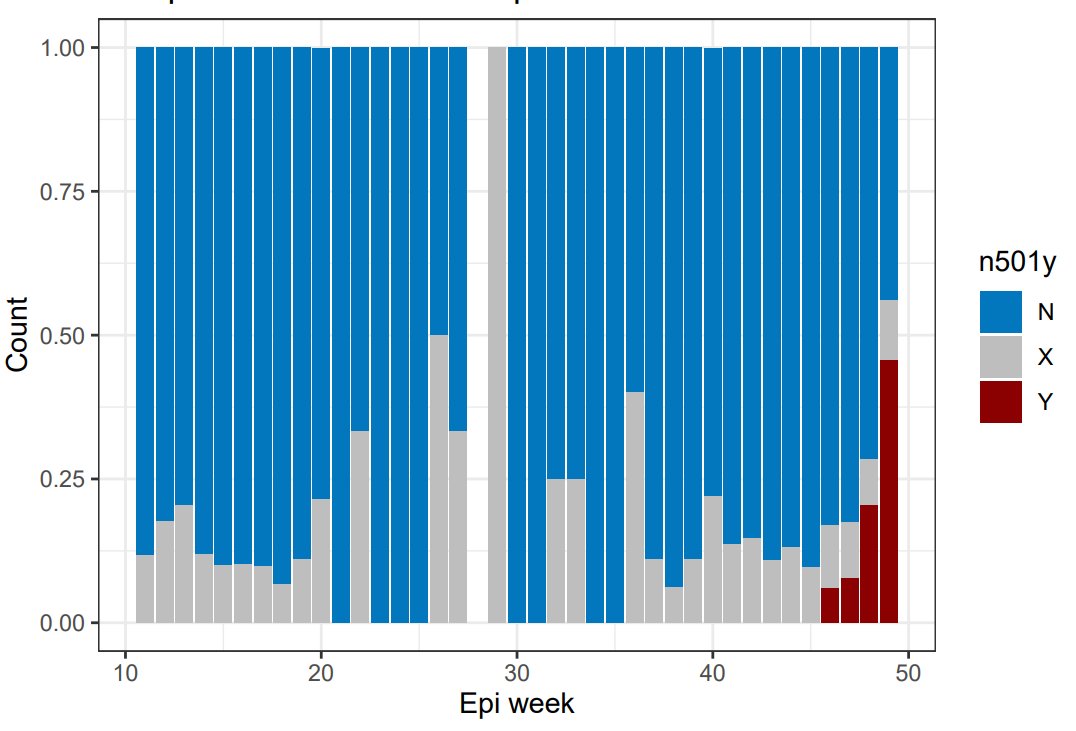
Microreact data over recent months, with rise of N501Y in Kent
re: this, Chris Whitty said at press conf that false negatives wouldn't be an issue
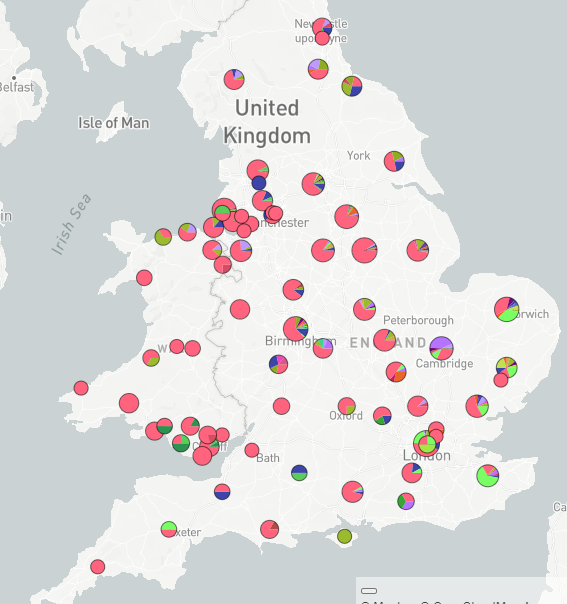
(clearer pies - but the relevant pie is Kent rather than the accidentally circled one)
A sense of the spread of this mutation. (This is a rough map of the UK: points represent genomes, with locations jittered and approximate) gist.github.com/theosanderson/…
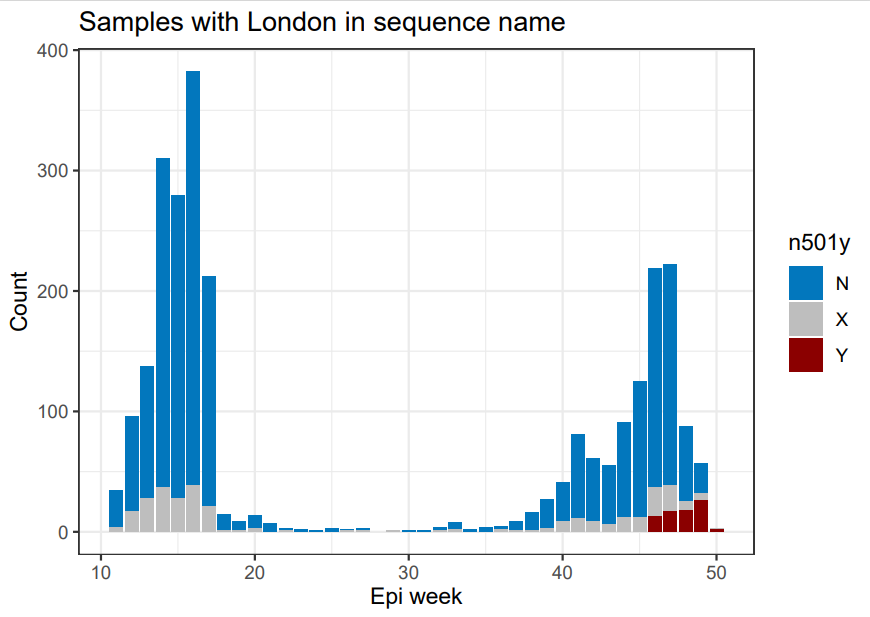
More data should go up on microreact tomorrow. The last mostly-complete week at present is 8 Nov - 14 Nov. Here's how London and Kent look there (lime green substantially up on prev. week). Let's see how 15 Nov - 21 Nov looks tomorrow. If these slices expand again that seems bad. 
(microreact didn't get pushed out today - I guessed it was on Wed because 9 Dec is in the current location name, but maybe it varies/appears a day later than the build)
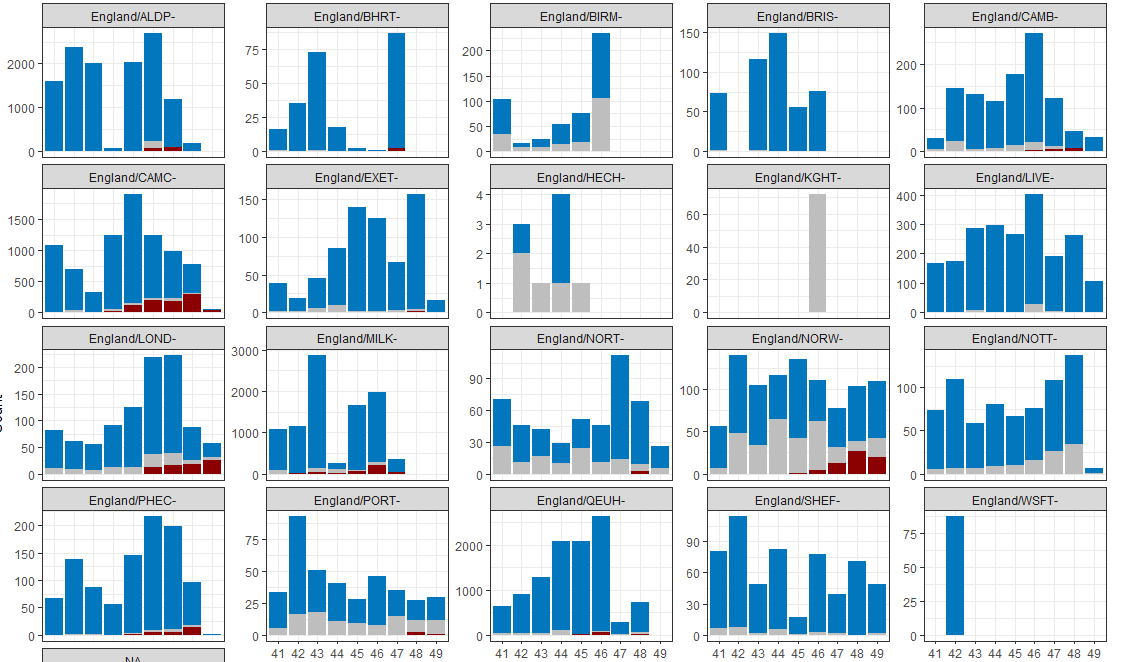
These annotations are about where the sequencing happened rather than where the sample was from, but there is correlation in broad geographical terms. LOND- London, CAMC = Cambridge Lighthouse, NORW - Norwich., MLK - Milton Keynes lighthouse. PHEC - PHE 
• • •
Missing some Tweet in this thread? You can try to
force a refresh