OK Koch heads... bring your hurt.
I’m glad you folks exist as we need people questioning everything.
However, I have found the “virus doesn’t exist” arguments unconvincing.
The virus isn’t the only cause of disease may have some ground.
I remain open to new papers on this.
I’m glad you folks exist as we need people questioning everything.
However, I have found the “virus doesn’t exist” arguments unconvincing.
The virus isn’t the only cause of disease may have some ground.
I remain open to new papers on this.

Set some rules.
This was established in 1884. Watson & Crick discovered DNA structure in 1953.
He did not contemplate unculturable organisms... the vast majority of organisms are not culture-able.
Viruses by definition cannot be cultured. They require a host cell to culture.
This was established in 1884. Watson & Crick discovered DNA structure in 1953.
He did not contemplate unculturable organisms... the vast majority of organisms are not culture-able.
Viruses by definition cannot be cultured. They require a host cell to culture.

This is where the debate gets dirty.
In order to culture the virus you need to find cells/organisms it can infect but not be a pathogen to those cells.
But Koch wants to demonstrate the pathogen creates symptoms of the disease which be counterproductive to culture.
In order to culture the virus you need to find cells/organisms it can infect but not be a pathogen to those cells.
But Koch wants to demonstrate the pathogen creates symptoms of the disease which be counterproductive to culture.
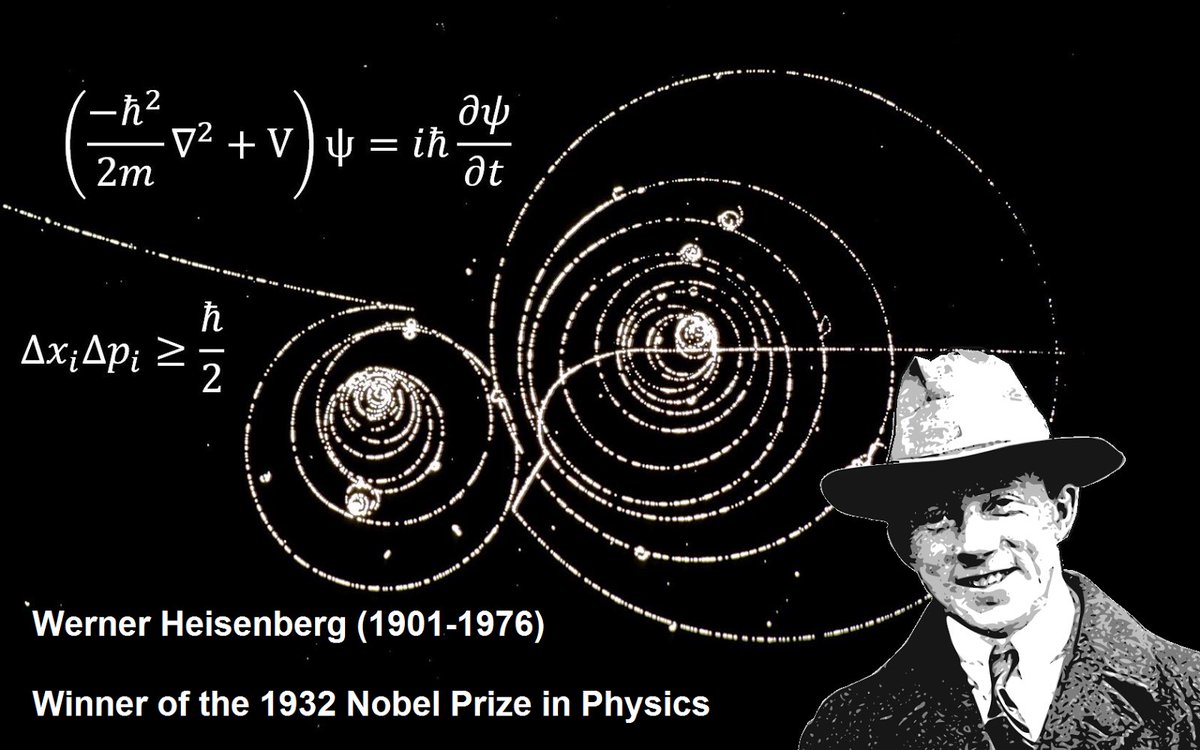
This is like the uncertainty principle.
The cell line/model organism you choose to culture the virus, can’t be extremely affected by the virus or the culture won’t succeed.
Therefore the vector for the disease is unlikely to be the best place to look for the disease symptoms.
The cell line/model organism you choose to culture the virus, can’t be extremely affected by the virus or the culture won’t succeed.
Therefore the vector for the disease is unlikely to be the best place to look for the disease symptoms.
As a result we have folks discounting Vero cell (monkey kidney cells) studies for viral isolation.
By this same logic, all animal models are thus insufficient as well.
Any argument that Vero cells are not identical to patients must also reject all animal models.
By this same logic, all animal models are thus insufficient as well.
Any argument that Vero cells are not identical to patients must also reject all animal models.
Medical ethics prevents us from infecting real patients with a virus suspected of pandemic potential.
This leaves us with Ex-vivo culture of sick and healthy patients samples.
Fairly invasive research. Needs IRB approval.
thelancet.com/journals/lanre…
This leaves us with Ex-vivo culture of sick and healthy patients samples.
Fairly invasive research. Needs IRB approval.
thelancet.com/journals/lanre…
This extracts epithelial cells from patients respiratory tract and cultures those cells as a model for viral infection and replication.
It has been done in C19 and I’m open to Koch Folks scrutinizing it’s short comings.
It has been done in C19 and I’m open to Koch Folks scrutinizing it’s short comings.
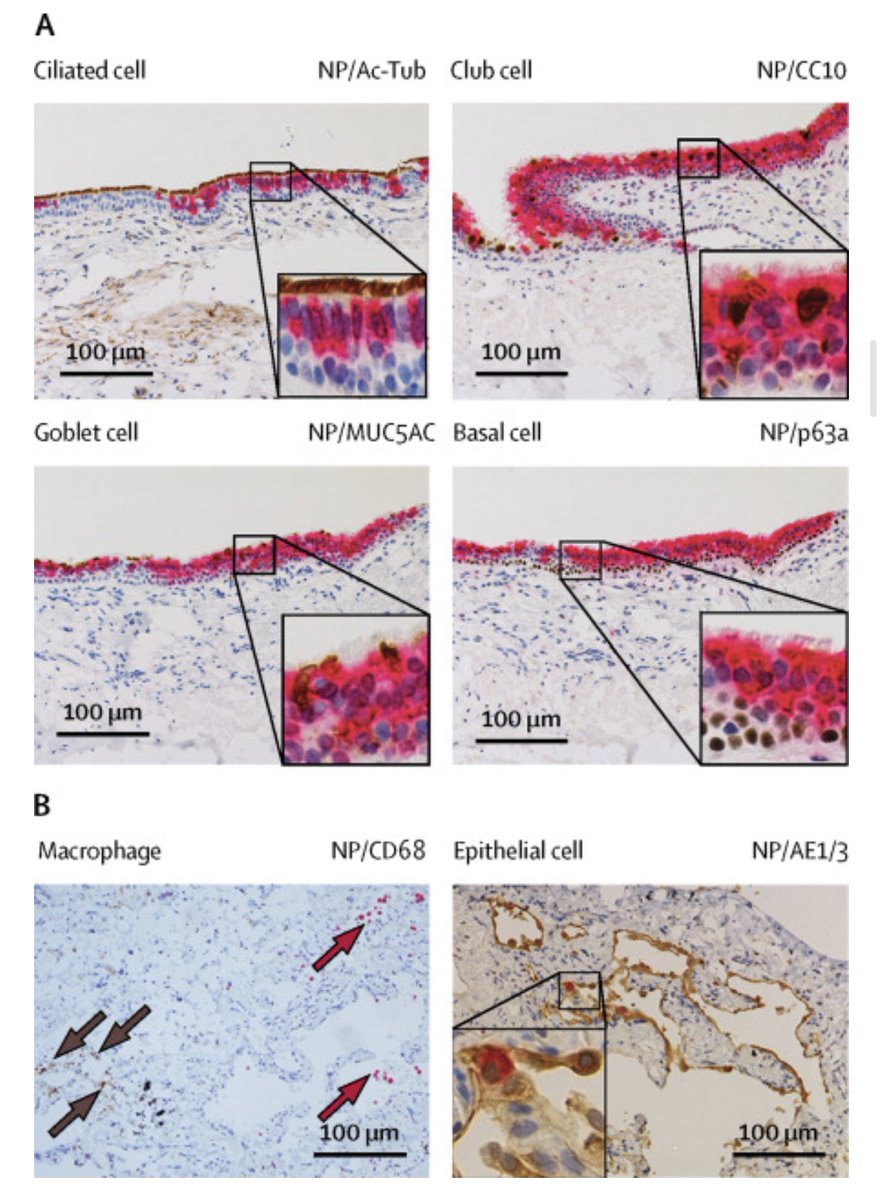
Ideally, we would have highly purified virus from the diseased and be able to put it into a human cell line and demonstrate replication and illness and re-isolate and confirm its the same.
Since we can’t infect humans, we resort to human cell lines.
Since we can’t infect humans, we resort to human cell lines.
We have new tools for understanding purity today.
We can sequence all RNA in a patient and see the complete virome present.
Based on the sequence we can predict proteins we should see with Mass spec after successful viral replication in a host.
biorxiv.org/content/10.110…
We can sequence all RNA in a patient and see the complete virome present.
Based on the sequence we can predict proteins we should see with Mass spec after successful viral replication in a host.
biorxiv.org/content/10.110…
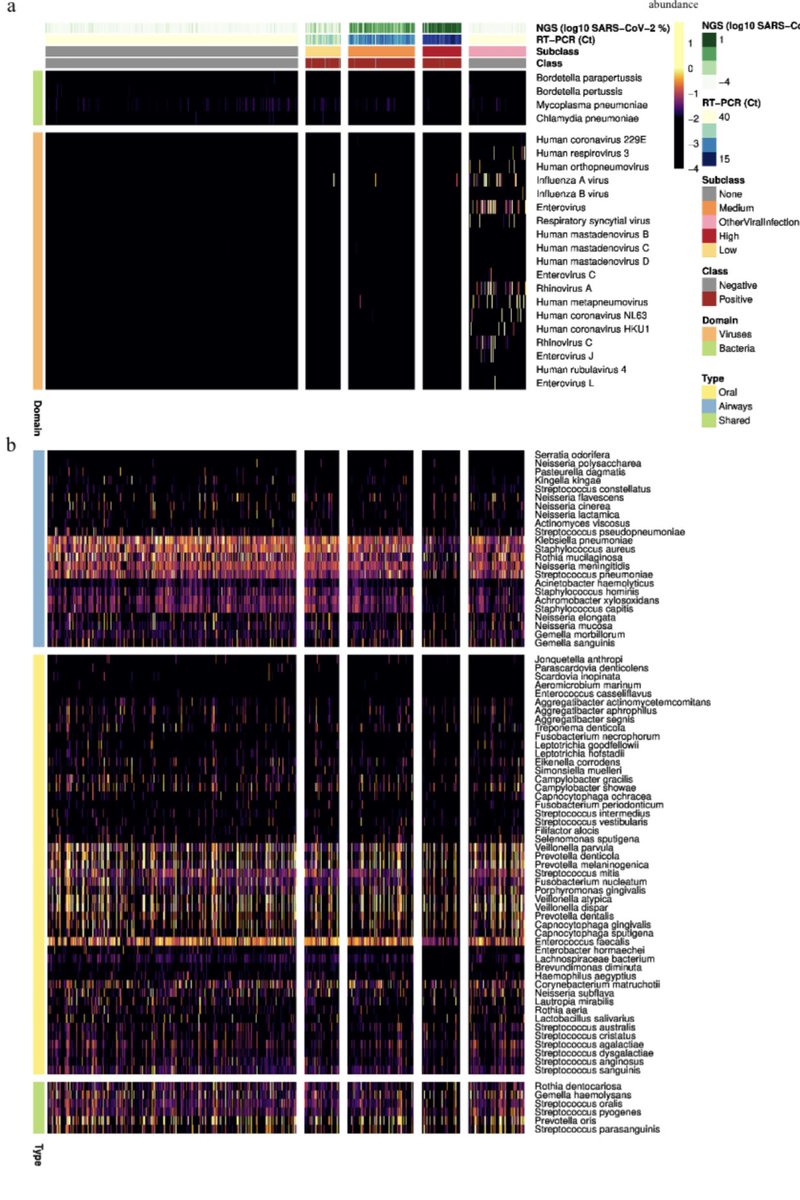
Masons lab goes on to build beautiful whole transcriptome pictures of each SARs patient.
Over 96% of the patients had C19 sequence in their Bronchial Lavage (BAFL). This is not Vero cell culture. Patient cells.
Whole SARs genomes from patients and it’s the dominant RNA present.
Over 96% of the patients had C19 sequence in their Bronchial Lavage (BAFL). This is not Vero cell culture. Patient cells.
Whole SARs genomes from patients and it’s the dominant RNA present.
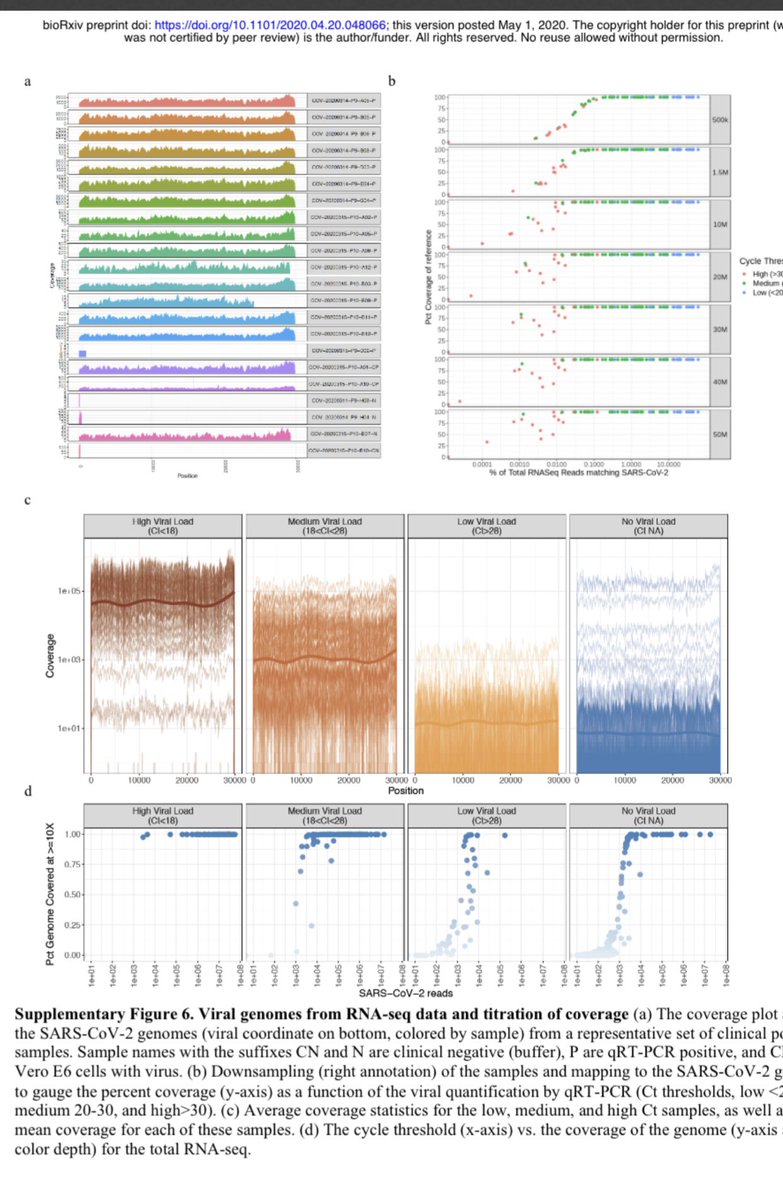
Some will say... you don’t know it’s the causative virus from this. Highest prevalence does not = causative agent.
But since we have whole transcriptomes we can see there is no other sensible hypothesis. No other viruses present.
The RNA has been isolated but not the virus.
But since we have whole transcriptomes we can see there is no other sensible hypothesis. No other viruses present.
The RNA has been isolated but not the virus.
On the flip side, images of fuzzy crowns on EM, I find unconvincing.
You can’t identify a virus with a photograph. All taxonomy is ultimately DNA/RNA based.
So how do we catch the virus in the act of infection?
Spatial transcriptomics-
This is the art of sequencing in situ.
You can’t identify a virus with a photograph. All taxonomy is ultimately DNA/RNA based.
So how do we catch the virus in the act of infection?
Spatial transcriptomics-
This is the art of sequencing in situ.
You infect cells with the purified virus and you perform spatial transcriptomics on those cells.
This is a new, explosive and exciting field that SOLiD sequencing is still being used for.
sciencemag.org/custom-publish…
This is a new, explosive and exciting field that SOLiD sequencing is still being used for.
sciencemag.org/custom-publish…
It’s worth understanding the technique.
It enables you to harvest RNA from cells and maintain their spatial coordinates in the cell with 10um and eventually 1um resolution.
It’s unlikely to get below the diffraction limit of light (250nm) in terms of resolution.
It enables you to harvest RNA from cells and maintain their spatial coordinates in the cell with 10um and eventually 1um resolution.
It’s unlikely to get below the diffraction limit of light (250nm) in terms of resolution.
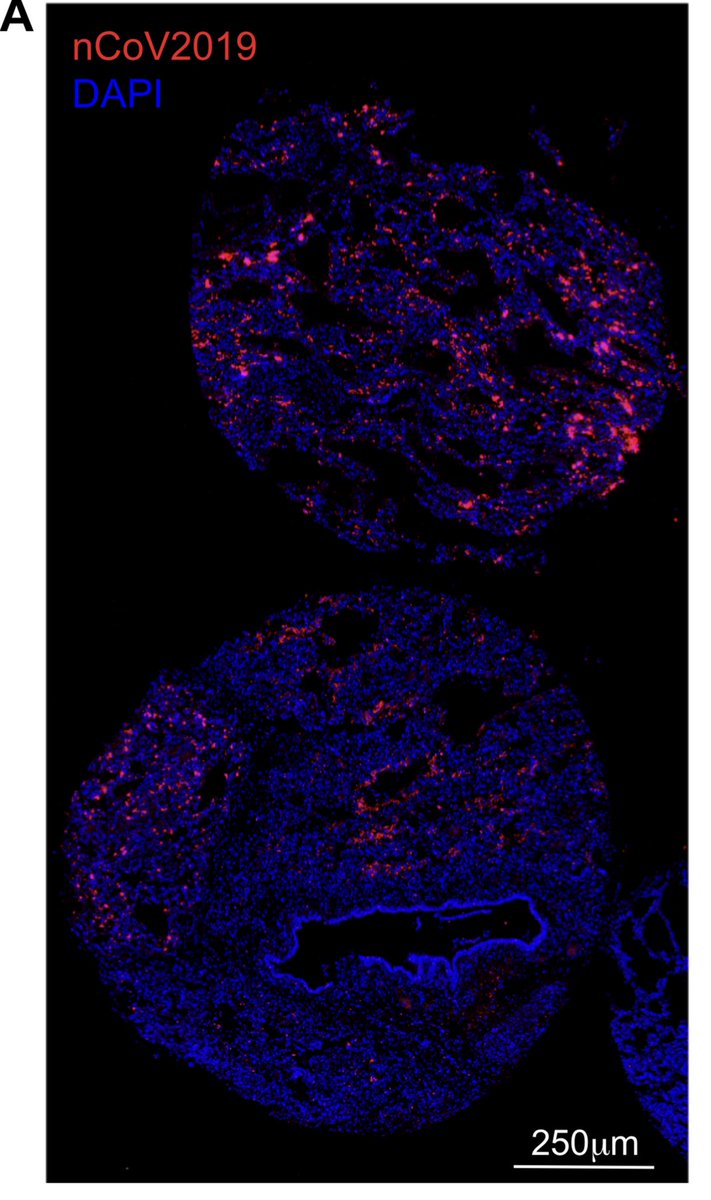
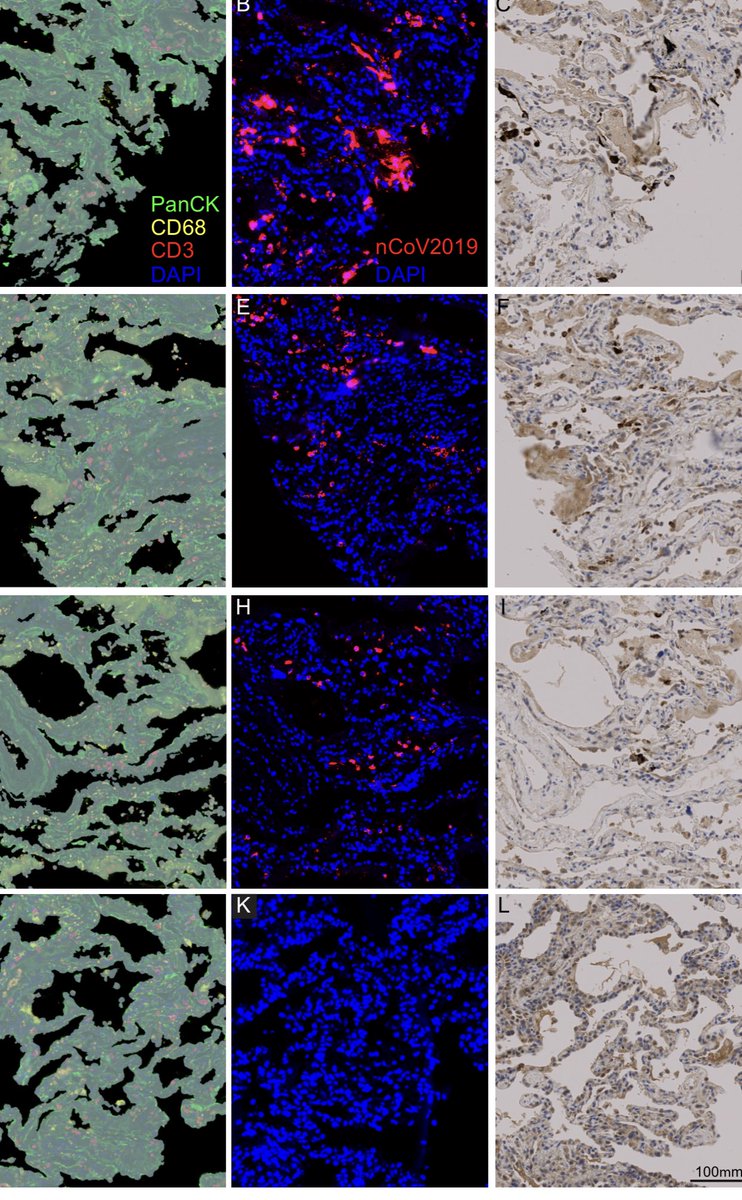
Here it deployed on C19 patient biopsies.
We have spike protein histochemistry co-localized with C19 RNA sequence in infected patients.
Not all biopsies succeeded. Lots of tissue heterogeneity but a good piece of the puzzle.
medrxiv.org/content/10.110…
We have spike protein histochemistry co-localized with C19 RNA sequence in infected patients.
Not all biopsies succeeded. Lots of tissue heterogeneity but a good piece of the puzzle.
medrxiv.org/content/10.110…
Now many are of the opinion that we need purified virus particle for the story to be complete.
I think this statement is stuck in 1884.
Viruses evolved from Viroids.
Viroids are just RNA. No capsid.
Small RNA alone can infect cells.
apsjournals.apsnet.org/doi/full/10.10…
I think this statement is stuck in 1884.
Viruses evolved from Viroids.
Viroids are just RNA. No capsid.
Small RNA alone can infect cells.
apsjournals.apsnet.org/doi/full/10.10…

So we have to keep our eyes on the code of the disease, not it’s wrapper.
The code dictates its proteins and identity. The wrapper is a non-specific, low information content camouflage that provides nice eye candy for journalists but provides little signature for function or ID.
The code dictates its proteins and identity. The wrapper is a non-specific, low information content camouflage that provides nice eye candy for journalists but provides little signature for function or ID.

So when people say, “you haven’t purified it”.
You can’t claim A causes B because you can’t assure me A is only A.
This is misunderstanding what Whole transcriptomes do.
When you sequence everything, you can then choose to synthesize the single RNA you think is causative.
You can’t claim A causes B because you can’t assure me A is only A.
This is misunderstanding what Whole transcriptomes do.
When you sequence everything, you can then choose to synthesize the single RNA you think is causative.

This is as pure as you can get. Even more pure than a cesium gradient isolation of the particles as the biological isolation will have biological contaminants not found in a synthesized from scratch genome.
We can also test the hypothesis. Knock out a gene and see what happens
We can also test the hypothesis. Knock out a gene and see what happens

4,000 times in one study.
But this won’t be pure enough for the Koch crowd.
pubmed.ncbi.nlm.nih.gov/34792434/
But this won’t be pure enough for the Koch crowd.
pubmed.ncbi.nlm.nih.gov/34792434/
The above study I did not think was ethical allowable but it appears I was wrong.
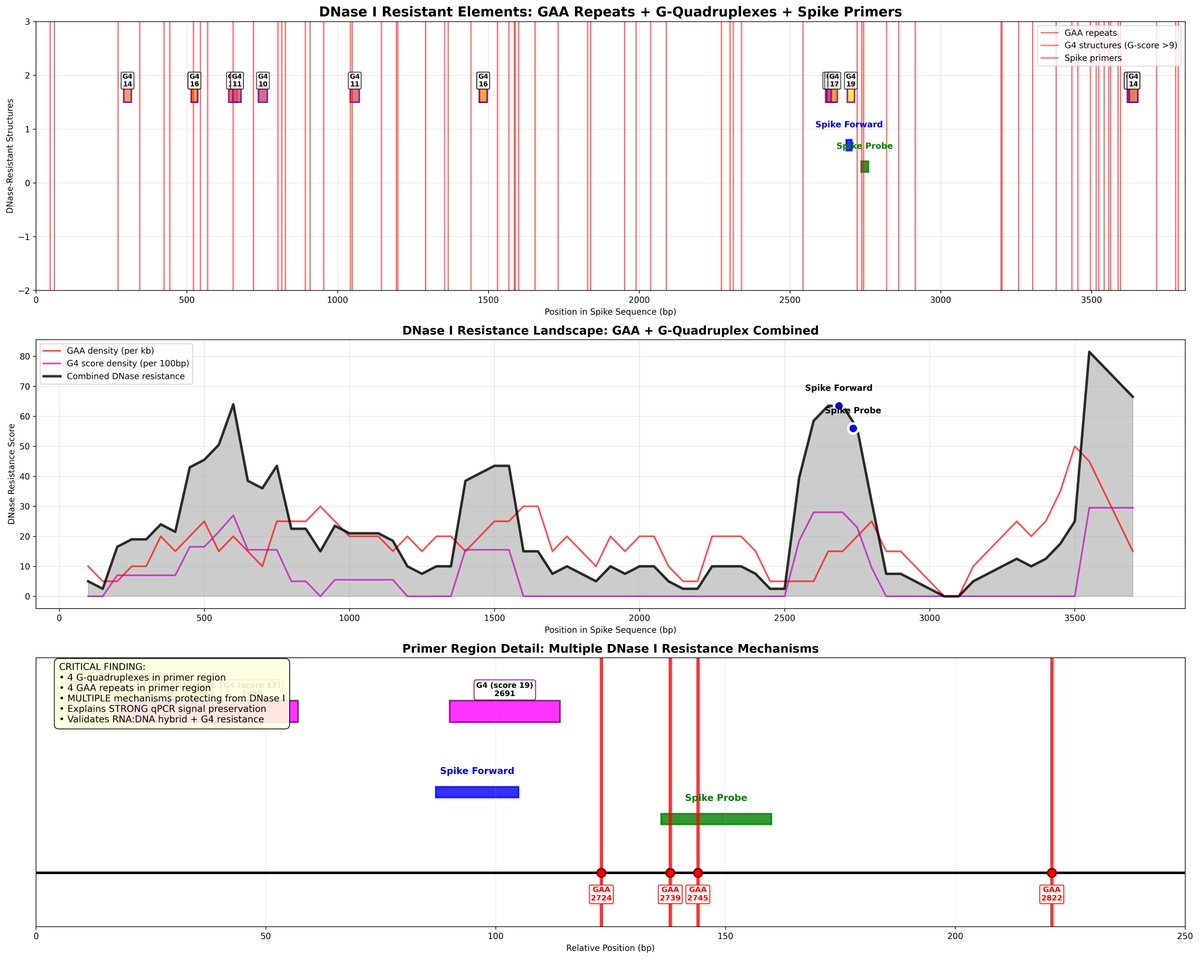
Important detail. After inoculation, they track the viral genomes growth through qPCR. You can see it climbing in copy number and decay over time. The increase is important as it cant be explained by the dilution of the inoculation into the host. The RNA sequence replicated. 

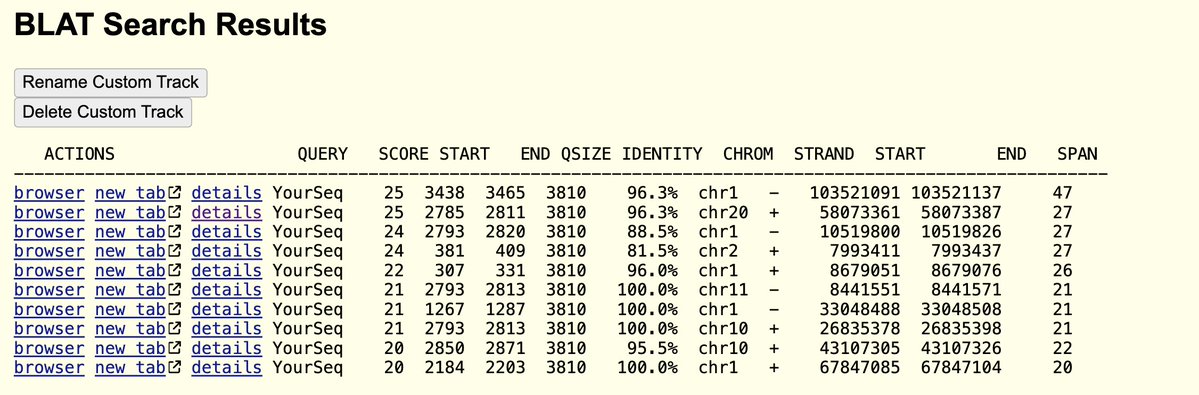
Now that the human genome has been fully sequenced…
There is no C19 sequence in it!
So where does this C19 RNA come from?
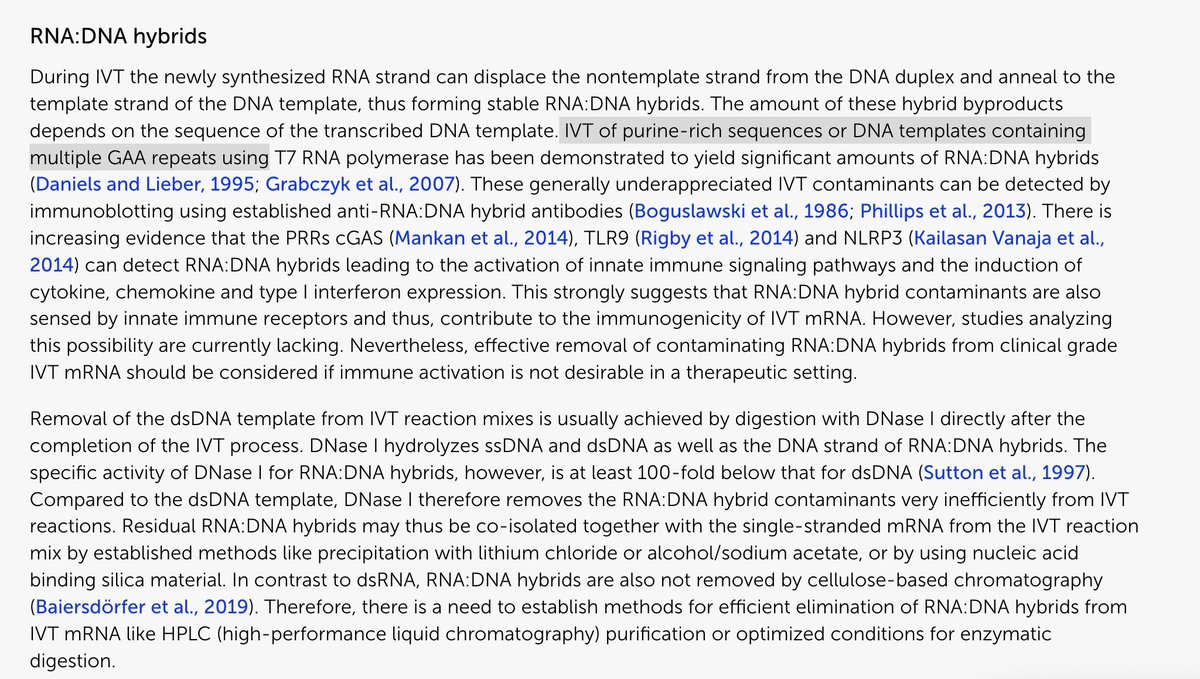
Mark Baileys excuse- “Barbara McClintock showed DNA shuffles”.
This is a mathematically illiterate response.
science.org/doi/10.1126/sc…
There is no C19 sequence in it!
So where does this C19 RNA come from?
Mark Baileys excuse- “Barbara McClintock showed DNA shuffles”.
This is a mathematically illiterate response.
science.org/doi/10.1126/sc…
Obelisks add to the support.
anandamide.substack.com/p/obelisks-and…
anandamide.substack.com/p/viroids-and-…
anandamide.substack.com/p/obelisks-and…
anandamide.substack.com/p/viroids-and-…
• • •
Missing some Tweet in this thread? You can try to
force a refresh