
What can we learn from sequencing (100% genetically identical?) monozygotic twin pairs?
Turns out, a lot, particularly on early embryo development. Here, deCODE deeply sequenced ~400 twin pairs, along with their children/parents when available.
1/10
nature.com/articles/s4158…
Turns out, a lot, particularly on early embryo development. Here, deCODE deeply sequenced ~400 twin pairs, along with their children/parents when available.
1/10
nature.com/articles/s4158…
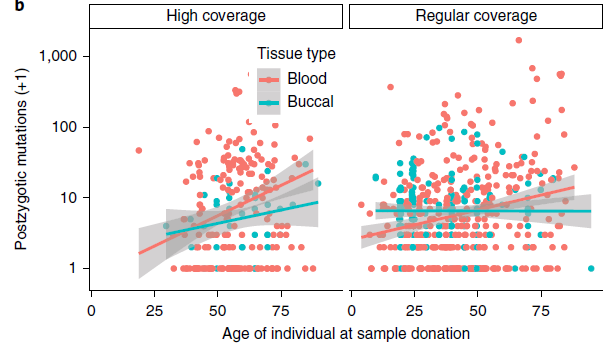
The authors found variability in the no. of postzygotic mutations (not in the twin's parents), e.g. 39 twin pairs differed by >100 mutations, 38 pairs did not differ at all.
The number of mutations increased with age, indicating that most of them accumulate through life.
2/10

The number of mutations increased with age, indicating that most of them accumulate through life.
2/10

More interesting are mutations that appear in a single twin + a child of that twin. These mutations must have occurred during early development, before the specification of the primordial germ cells, as they appear in both soma (blood/cheeks) and germline (children).
3/10
3/10

They found that each twin had ~1.3 such mutations (among the total ~60 transmitted to their children). Since only half the mutations are transmitted, twins differ in 4*1.3=5.2 early developmental mutations. The number was highly variable across families.
4/10
4/10
In ~15% of families, a twin transmitted many more (3.5) early mutations, and these mutations also had variant allele frequency in blood near 50%. Those individuals may have been derived from a single cell, which accumulated mutations before diverging from the other twin.
5/10
5/10
Looking at early mutations present in both twins, the authors found high variability across families in how similar allele frequencies are between the twins. This includes some extreme cases, as in the pictured pedigrees.
6/10
6/10

These results overall imply high randomness in the number/diversity of cells that generate each twin. Differences between twins could be due to one twin arising from just a single/few cells, or from extensive cell death in that twin quickly after twinning.
7/10
7/10
Further, they found that somatic and germline allele frequencies of early mutations are strongly correlated. (Germline frequency is estimated by the probability of presence in additional children). Thus, the same founder cells give rise to both germ/somatic cells.
8/10
8/10

Finally, they found more C>T/indels and less T>C/CpG>TpG mutations during early cell divisions. This is interpreted as (1) lower activity of transcription-coupled DNA repair and (2) lower global CpG methylation during early development.
9/10
9/10

I find it truly amazing how much can be learned about early embryo development based on "indirect" evidence of genome sequences of living twins and their family members. This paper is really a tour de force!
10/10
10/10
• • •
Missing some Tweet in this thread? You can try to
force a refresh


