🙏🏼 for the follows.
My goal is to simply share what I know about genomics with the public when it matters most.
I rarely get technical refutations but embrace the errors so as to correct.
The best they seem to be able to fire back is ad hominens and associative fallacies
My goal is to simply share what I know about genomics with the public when it matters most.
I rarely get technical refutations but embrace the errors so as to correct.
The best they seem to be able to fire back is ad hominens and associative fallacies
https://twitter.com/Gurdur/status/1347653581269331969
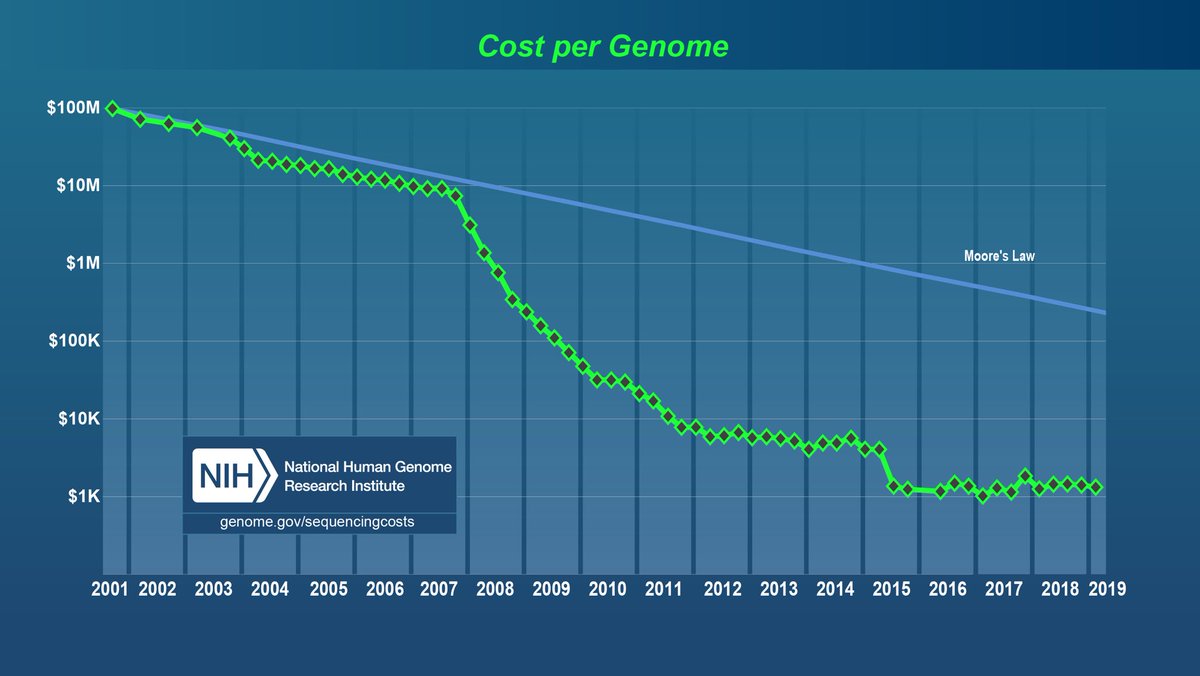
From 1996 to 2000 I worked tirelessly to get as much as human genome public as possible.
Our team then built a DNA sequencer that dropped the price of DNA sequencer several orders of magnitude.
Published open access.
genome.cshlp.org/content/19/9/1…
Our team then built a DNA sequencer that dropped the price of DNA sequencer several orders of magnitude.
Published open access.
genome.cshlp.org/content/19/9/1…
In 2000 we started a genomic company called Agencourt.
It became the largest commercial seq shop at the time.
Turned profitable in 18 months from start-up.
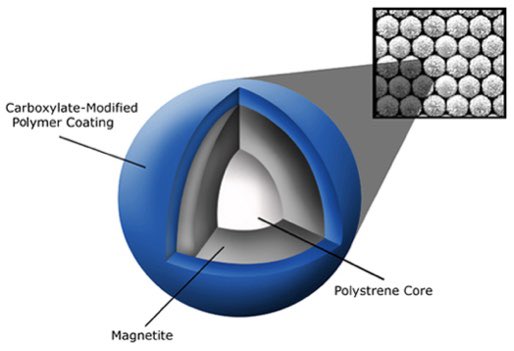
We also built DNA and RNA purification kits for viruses using SPRI
Some of these kits are in use today for C19 prep.
It became the largest commercial seq shop at the time.
Turned profitable in 18 months from start-up.
We also built DNA and RNA purification kits for viruses using SPRI
Some of these kits are in use today for C19 prep.
We also became one of 5 NHGRI genome centers along with MIT, Baylor, WashU, and Venter.
We built it in the burbs. Had low overhead and focused on plasmid whole genome shotgun and Fosmids.
Our niche play made us the cheapest center for these protocols.
We built it in the burbs. Had low overhead and focused on plasmid whole genome shotgun and Fosmids.
Our niche play made us the cheapest center for these protocols.
Beckman Coulter acquired the 120 person company and spun out Agencourt Personal Genomics which build the SOLiD sequencer and was acquired a year later as a 19 person start-up.
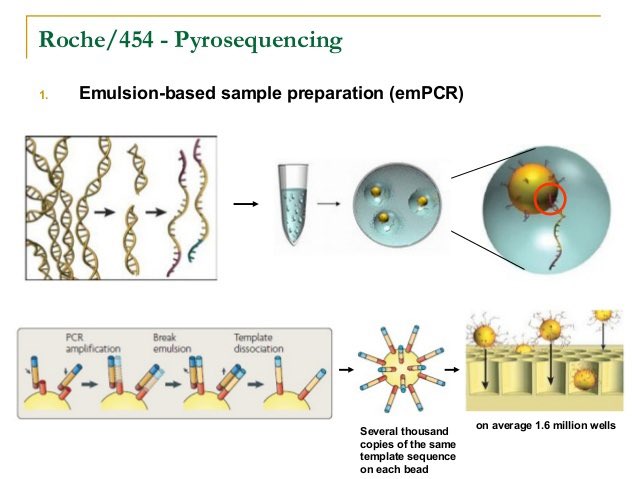
SOLIiD was based on a several revolutionary ideas.
Emulation PCR from JHU


SOLIiD was based on a several revolutionary ideas.
Emulation PCR from JHU
https://x.com/kevin_mckernan/status/1333220058831708163?s=21
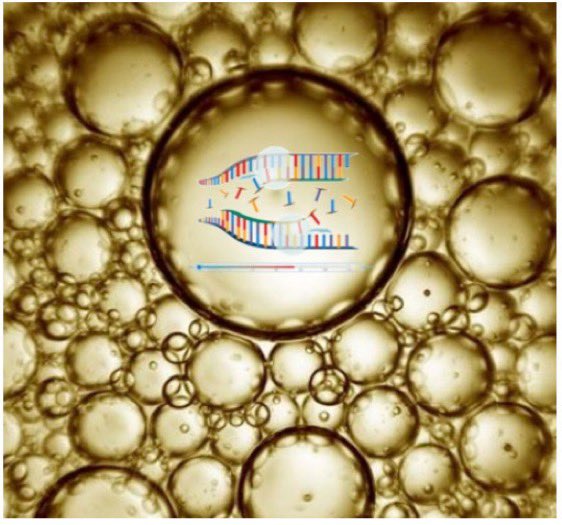
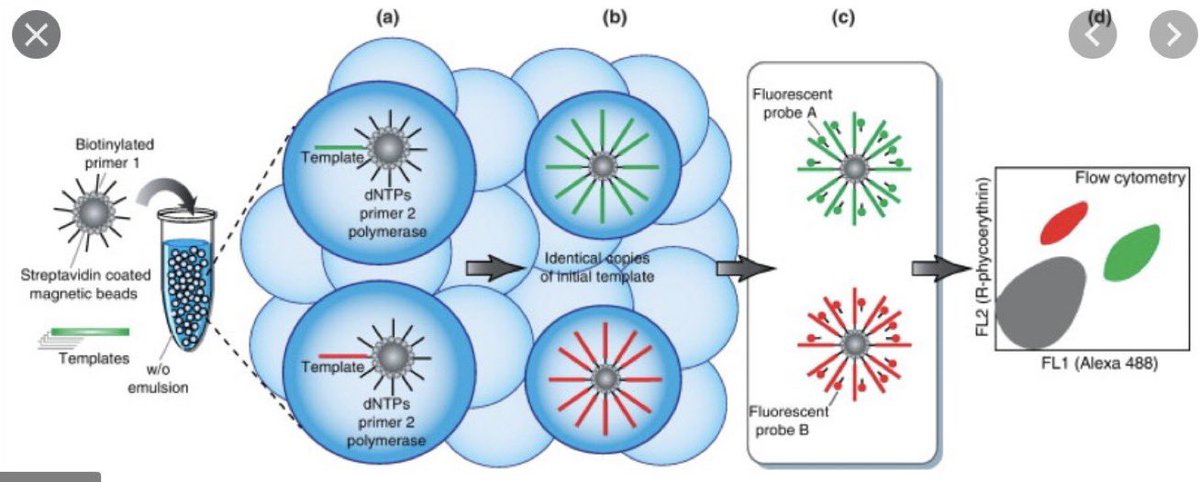
This enabled billions of individual 30um diameter PCR reactions to be performed in a single reaction vessel.
We’d put a 1um magnetic bead in each bubble that had one of the 2 PCR primers on it and we could get DNA to amplify to the bead and coat it like a tennis ball w/DNA.
We’d put a 1um magnetic bead in each bubble that had one of the 2 PCR primers on it and we could get DNA to amplify to the bead and coat it like a tennis ball w/DNA.
Now, if you put a low concentration of library of 🧬 that had sequence that matched the Bead primer and a second solution primer you could amplify a single molecule to bead and make 100,000 copies of the DNA on the magnetic particle.
Now each bead could be sequenced.
Now each bead could be sequenced.
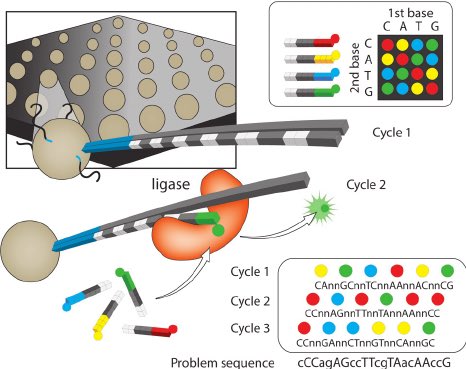
I bring this up because it made our team understand PCR at a single molecule level and at a depth no one had ever seen before.
We needed a deep understanding of Oligo synthesis and artifacts that impacted massively parallel single molecule sensitive PCR at scales never seen b4.
We needed a deep understanding of Oligo synthesis and artifacts that impacted massively parallel single molecule sensitive PCR at scales never seen b4.
This company was Acquired by Applied Biosystems which was then acquired by Life Tech and then Thermo.
Thermo is one the largest picks and shovels companies in PCR testing.
But it was time to give back.
We were sequencing cancer biopsies now...
Thermo is one the largest picks and shovels companies in PCR testing.
But it was time to give back.
We were sequencing cancer biopsies now...
We could now sequence human genomes for under $5K and that meant tumor sequencing could help personalize cancer therapy.
More work with JHU and SOLiD decorated the cover of Science T Med.
stm.sciencemag.org/content/2/20/2…
More work with JHU and SOLiD decorated the cover of Science T Med.
stm.sciencemag.org/content/2/20/2…

We could personalize but the drug choices were splitting hairs on poison.
Cannabinoids caught my attention.
It’s genome wasnt sequenced yet.
July 2011- flew to Amsterdam.
Purified DNA in a hotel room.
Sequence assembly public Aug 18.

Cannabinoids caught my attention.
It’s genome wasnt sequenced yet.
July 2011- flew to Amsterdam.
Purified DNA in a hotel room.
Sequence assembly public Aug 18.


At this turn in my career, I started to wise up to the scientific censorship and ridicule of certain fields of study.
I became an open source vigilante and became enthralled with censorship resistant Blockchains.
Published some clinical PCR methods to weaken gene patents.
I became an open source vigilante and became enthralled with censorship resistant Blockchains.
Published some clinical PCR methods to weaken gene patents.

We began thinking about how distributed consensus protocols could enhance scientific funding and transparency in Peer Review.
Ran another Cannabis genome project this way.
60 days from funding to public upload.
The THC/CBD+ Jamaican Lion genome.
Ran another Cannabis genome project this way.
60 days from funding to public upload.
The THC/CBD+ Jamaican Lion genome.
We decided to crack the stigma on the field and organized the CannMed conference at Harvard Medical School.
@CannMedEvents
Videos cover diverse topics from Cancer, Plant pathogens, tissue culture, FDA trials, extraction chemistry and genomics.
Public
cannmedevents.com/cannmed-video-…
@CannMedEvents
Videos cover diverse topics from Cancer, Plant pathogens, tissue culture, FDA trials, extraction chemistry and genomics.
Public
cannmedevents.com/cannmed-video-…
We have recently sequenced and put public Kratom and Psilocybe cubensis (magic mushrooms) as these genomes make natural break through therapies and we need these genomes in the public domain.
Public Psilocybe Cubensis Genome browser.
Public Psilocybe Cubensis Genome browser.
https://twitter.com/Kevin_McKernan/status/1330193833834012672
Why am I an open source zealot?
I realize innovation is a team sport and it’s rare all the team member are in your location.
Public data floats all boats. It accelerates innovation providing more opportunity to nimble and energetic curiosity.
Data in one mind is a waste.
I realize innovation is a team sport and it’s rare all the team member are in your location.
Public data floats all boats. It accelerates innovation providing more opportunity to nimble and energetic curiosity.
Data in one mind is a waste.
This also means I’ve become impatient with centralized and slow peer review as its prone to censorship and poor quality.
Performed anon and with conflicts ....and this how we seed fake news.
Performed anon and with conflicts ....and this how we seed fake news.
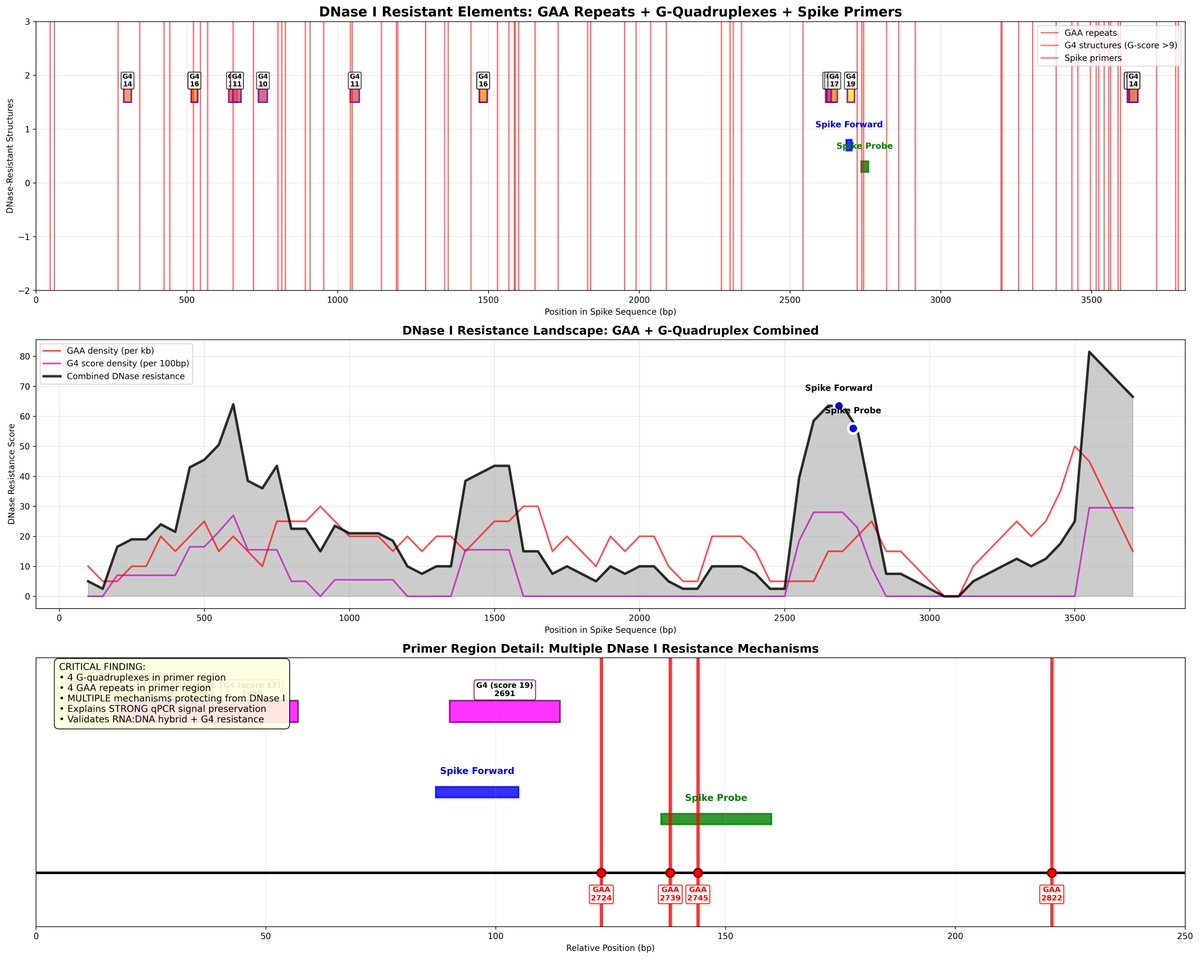
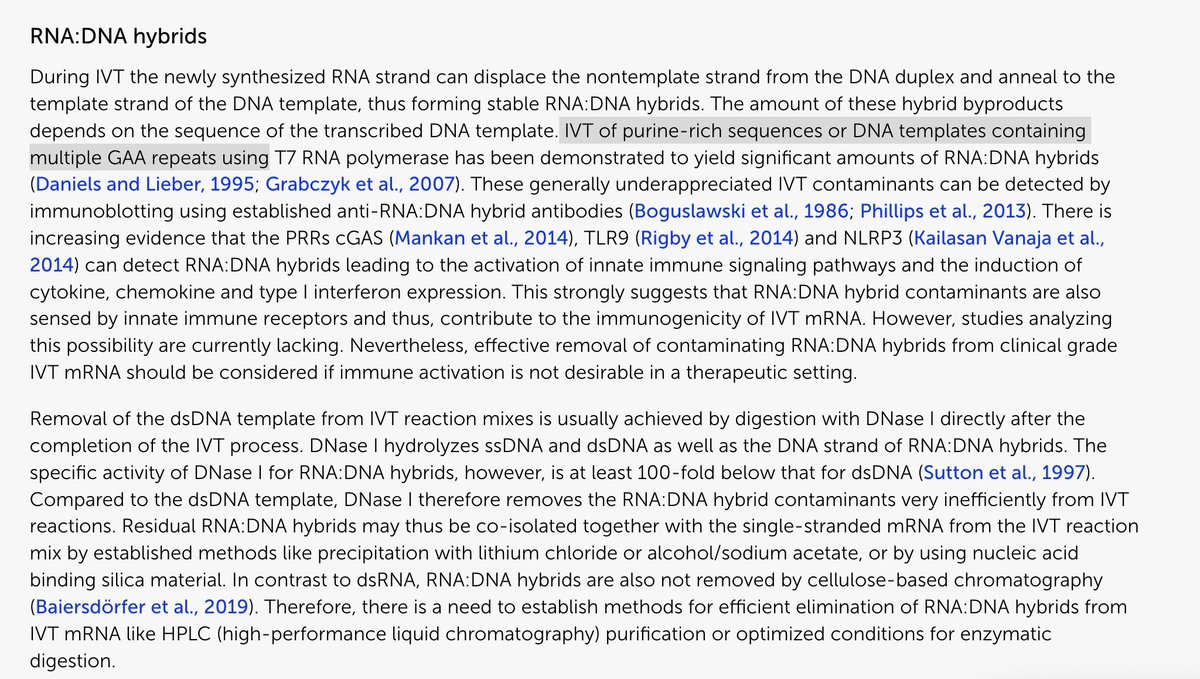
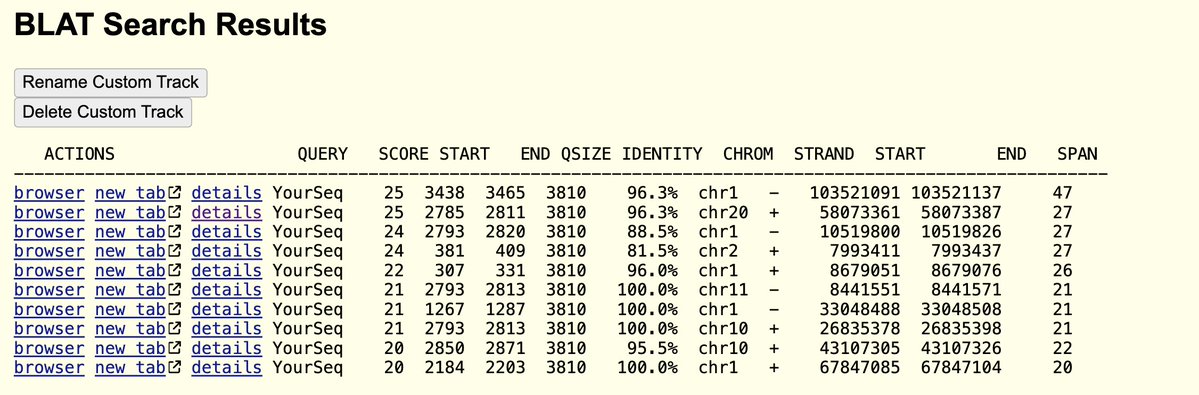
I think I’ve earned the public’s trust on being able to view in on the peer review of a PCR protocol that is currently choking the world.
Yet the wing of science communicators and merchants of despair can’t defend these critiques.
They slander.
Who do you believe?
Yet the wing of science communicators and merchants of despair can’t defend these critiques.
They slander.
Who do you believe?
• • •
Missing some Tweet in this thread? You can try to
force a refresh