After ~10 months of relative quiescence we've started to see some striking evolution of SARS-CoV-2 with a repeated evolutionary pattern in the SARS-CoV-2 variants of concern emerging from the UK, South Africa and Brazil. 1/19
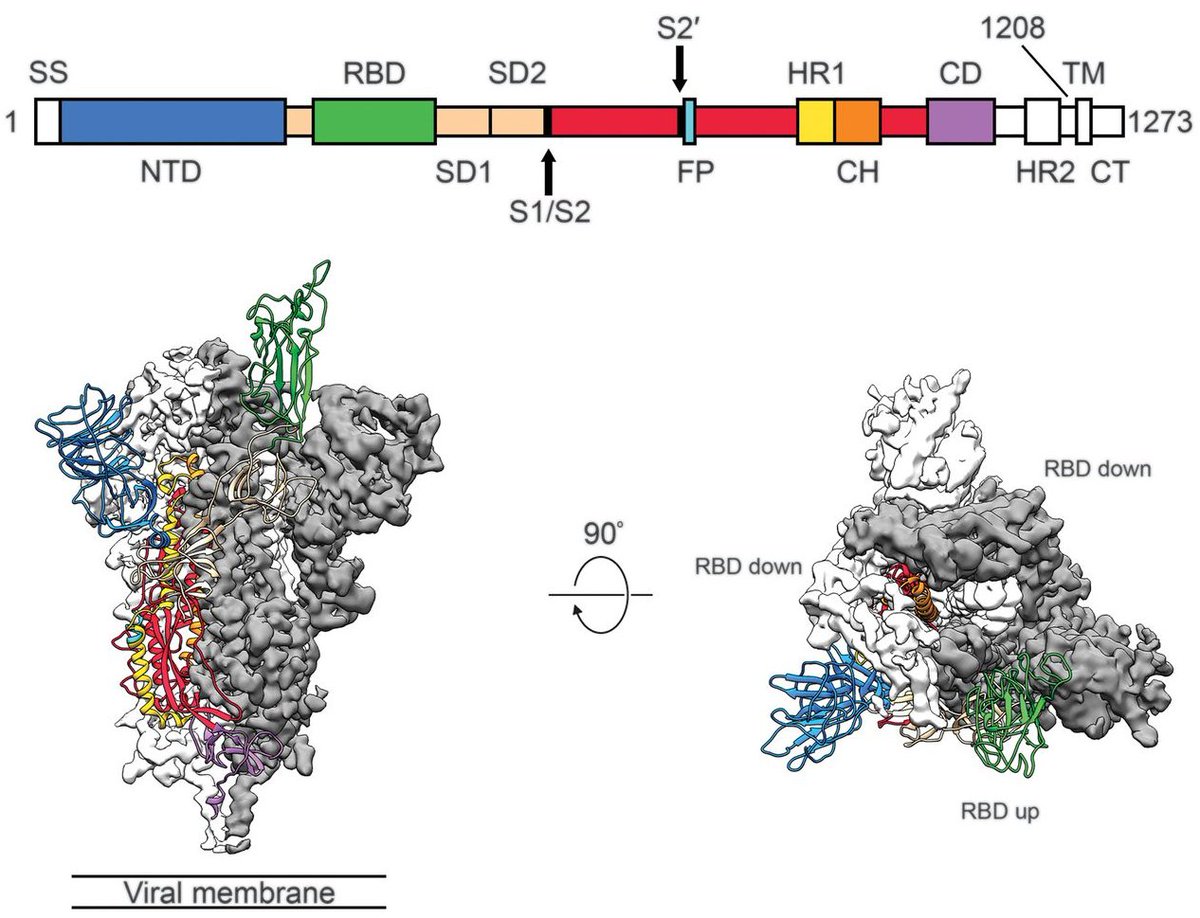
In SARS-CoV-2, the viral spike protein and in particular the receptor binding domain (RBD) is a locus for important viral evolution and is the primary target for the human immune response (figure from science.sciencemag.org/content/367/64…). 2/19 
There had been little evolution in the RBD until ~Oct 2020 when we saw RBD mutations start to spread. Perhaps chief among mutations of interest are E484K and N501Y which mutate nearby sites in the RBD. The evolution of these sites can be seen here: nextstrain.org/ncov/global?c=…. 3/19 
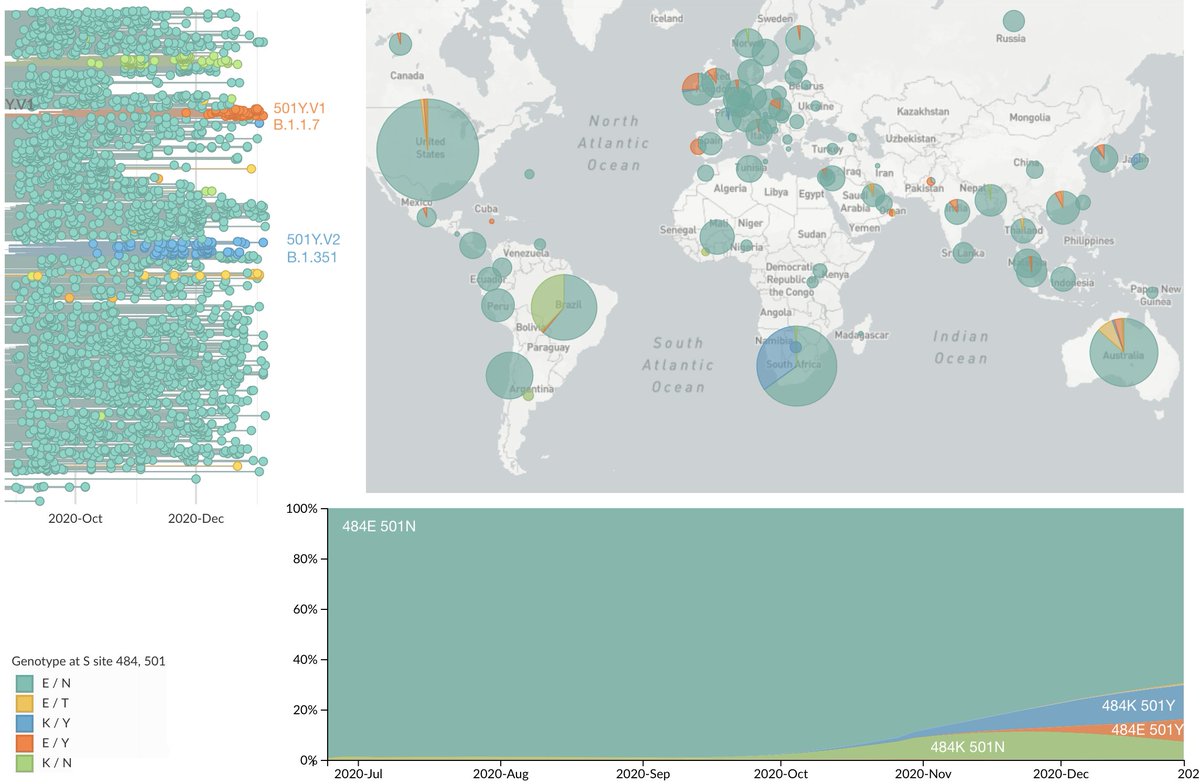
E484K and N501Y have occurred repeatedly on the evolutionary tree of the virus representing convergent evolution. Many 484K and 501Y instances are due to the three variants of concern, but there are other 484K and 501Y viruses that are not due to these variants. 4/19
The variant emerging in the UK (@nextstrain clade 20I/501Y.V1, cov-lineages.org lineage B.1.1.7, described at virological.org/t/preliminary-…) possesses 8 mutations in spike, a 3 amino acid deletion in ORF1ab and an ORF8 knockout mutation. nextstrain.org/ncov/europe?br… 5/19 
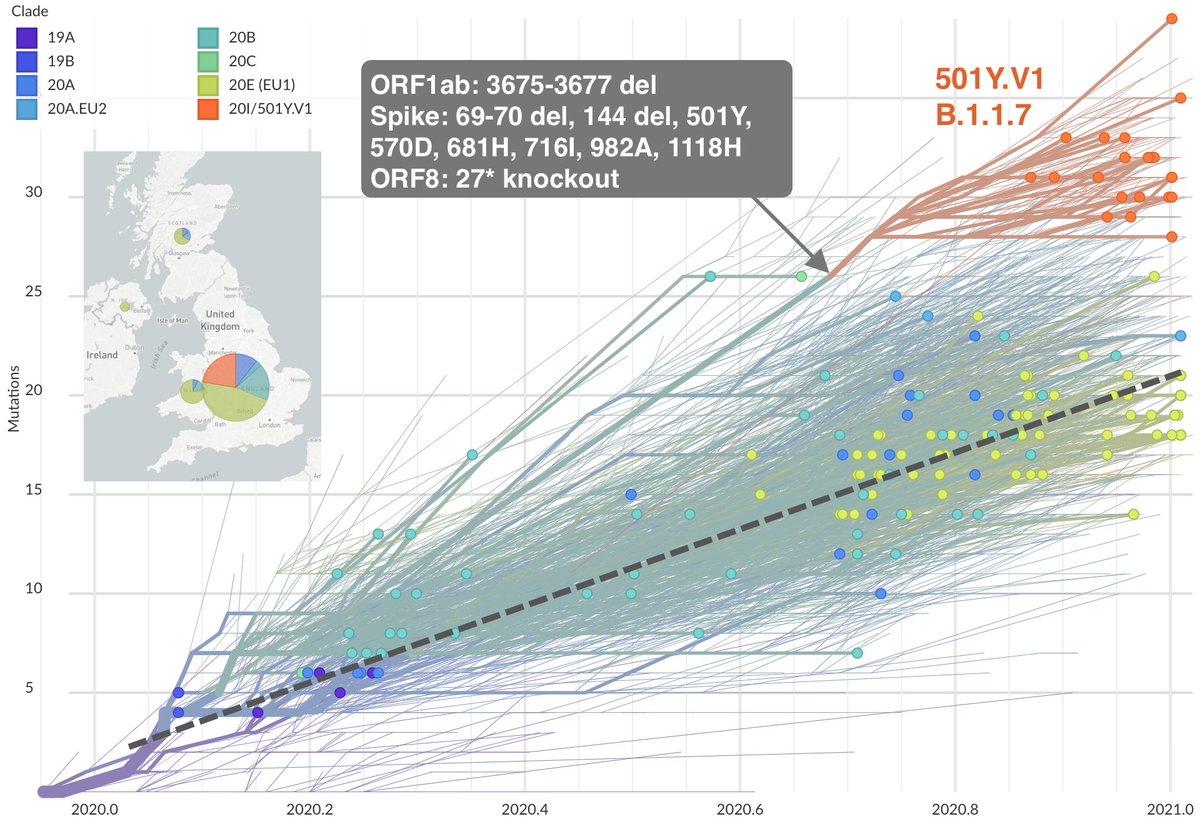
The above figure is a "root-to-tip" plot showing number of mutations in sampled viruses against date of collection. The dashed line represents average accumulation of mutations of about 1 mutation every 2 weeks. The 501Y.V1 variant stands out here with additional mutations. 6/19
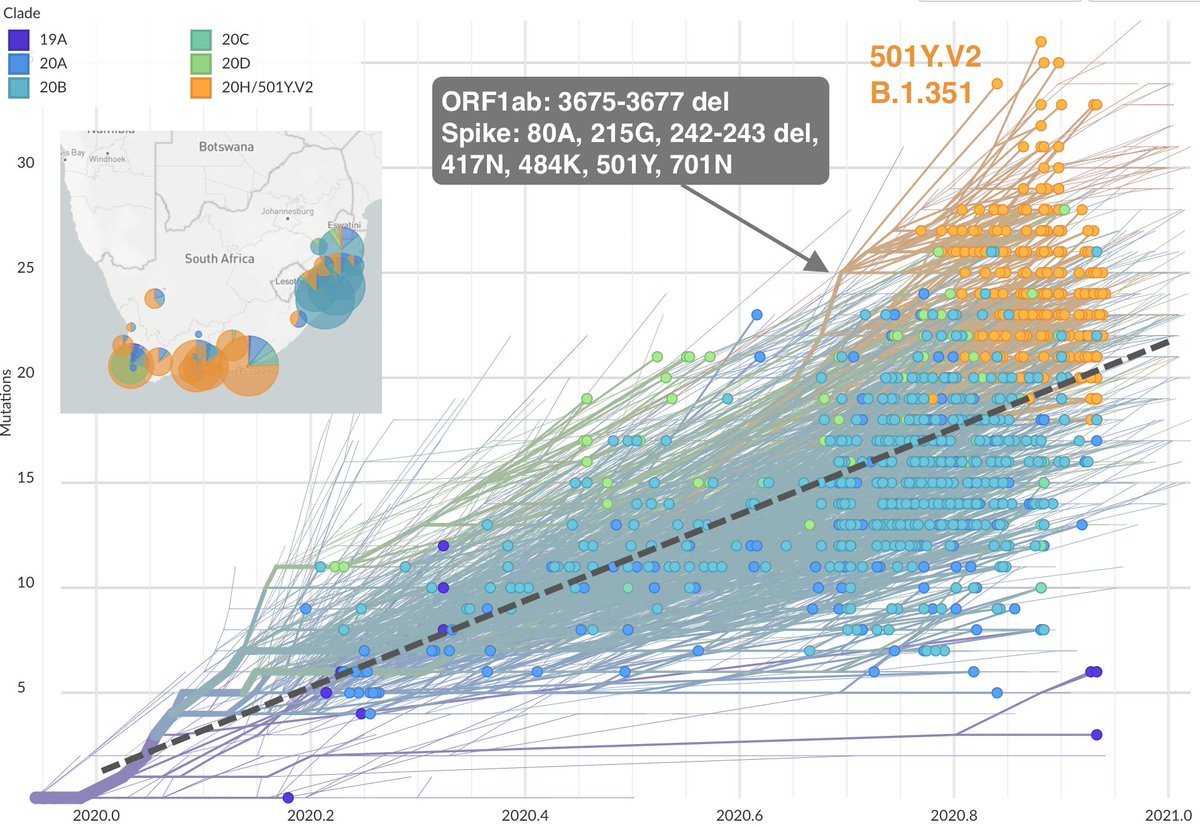
The variant emerging in South Africa (@nextstrain clade 20H/501Y.V2, cov-lineages.org lineage B.1.351, described at medrxiv.org/content/10.110…) has 7 mutations in spike as well as the same 3 amino acid deletion in ORF1ab. nextstrain.org/ncov/africa?br… 7/19 
The variant emerging in Brazil (@nextstrain clade 20J/501Y.V3, cov-lineages.org lineage P.1, described at virological.org/t/genomic-char…) has 10 mutations in spike as well as the same 3 amino acid deletion in ORF1ab. nextstrain.org/ncov/south-ame… 8/19 
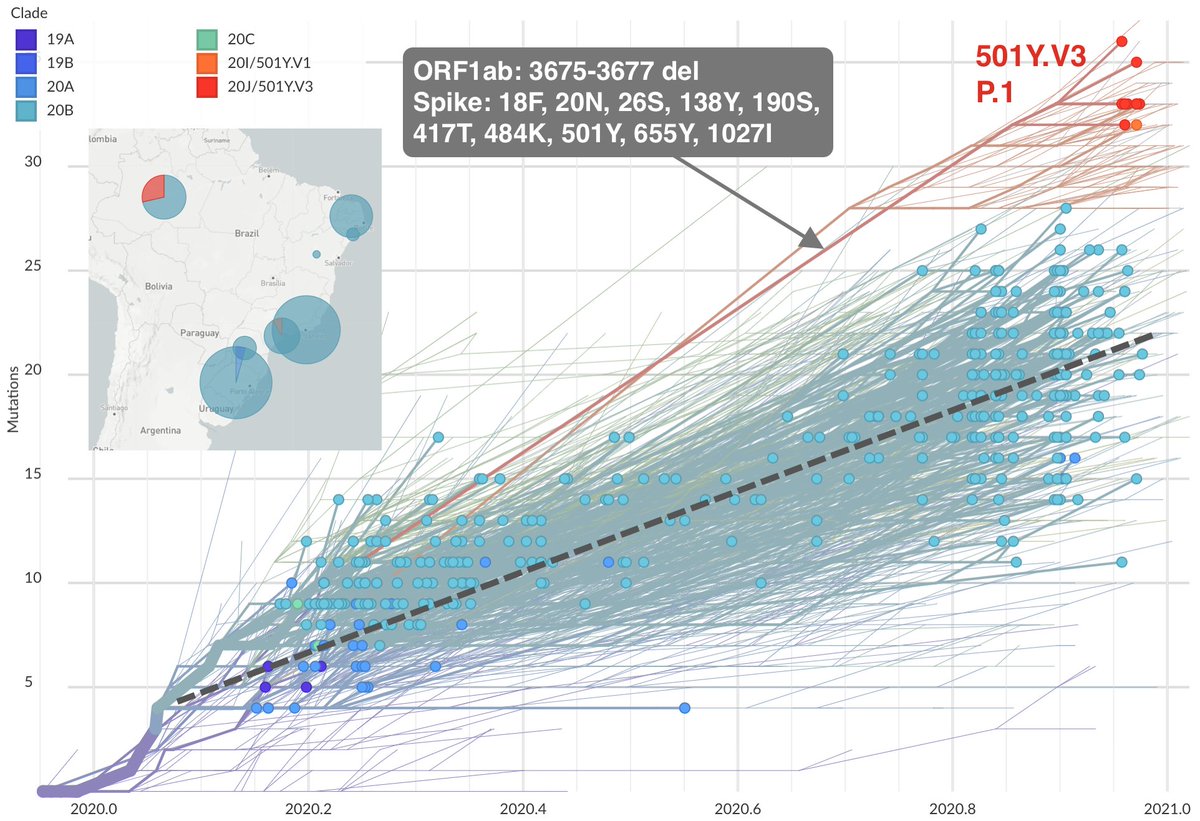
Like 501Y.V1, both 501Y.V2 and 501Y.V3 also appear to possess "extra" mutations beyond what's seen in most circulating viruses. And importantly, these variant viruses show repeated convergent evolution including spike sites 417, 484, 501 and the ORF1ab deletion. 9/19
Extra protein coding mutations localized to spike alongside convergent evolution are hallmarks of adaptive evolution. So, what's going on here? 10/19
My (highly speculative!) hypothesis is that the emergence of these variant viruses arises in cases of chronic infection during which the immune system places great pressure on the virus to escape immunity and the virus does so by getting really good at getting into cells. 11/19
This results in the observed increased viral load (medrxiv.org/content/10.110…) and greater transmissibility (gov.uk/government/pub…) of 501Y.V1 / B.1.1.7, but importantly this is not due to population-level selection for antigenic drift or faster transmission. 12/19
Instead it's a knock-on effect of the reactor of evolution occurring in rare chronic infections and then spillover from these infections into the greater population. 13/19
This hypothesis of increased binding and cell entry as mechanism for immune evasion was expressed previously in the context of seasonal influenza by @SCOTTeHENSLEY in science.sciencemag.org/content/326/59…. 14/19
Hints that these emergence events are related to chronic infections come from multiple sources. First off, we have direct sequencing of viruses from chronic infections that show rapid evolution (figure from @binachoimd et al nejm.org/doi/full/10.10…). 15/19 
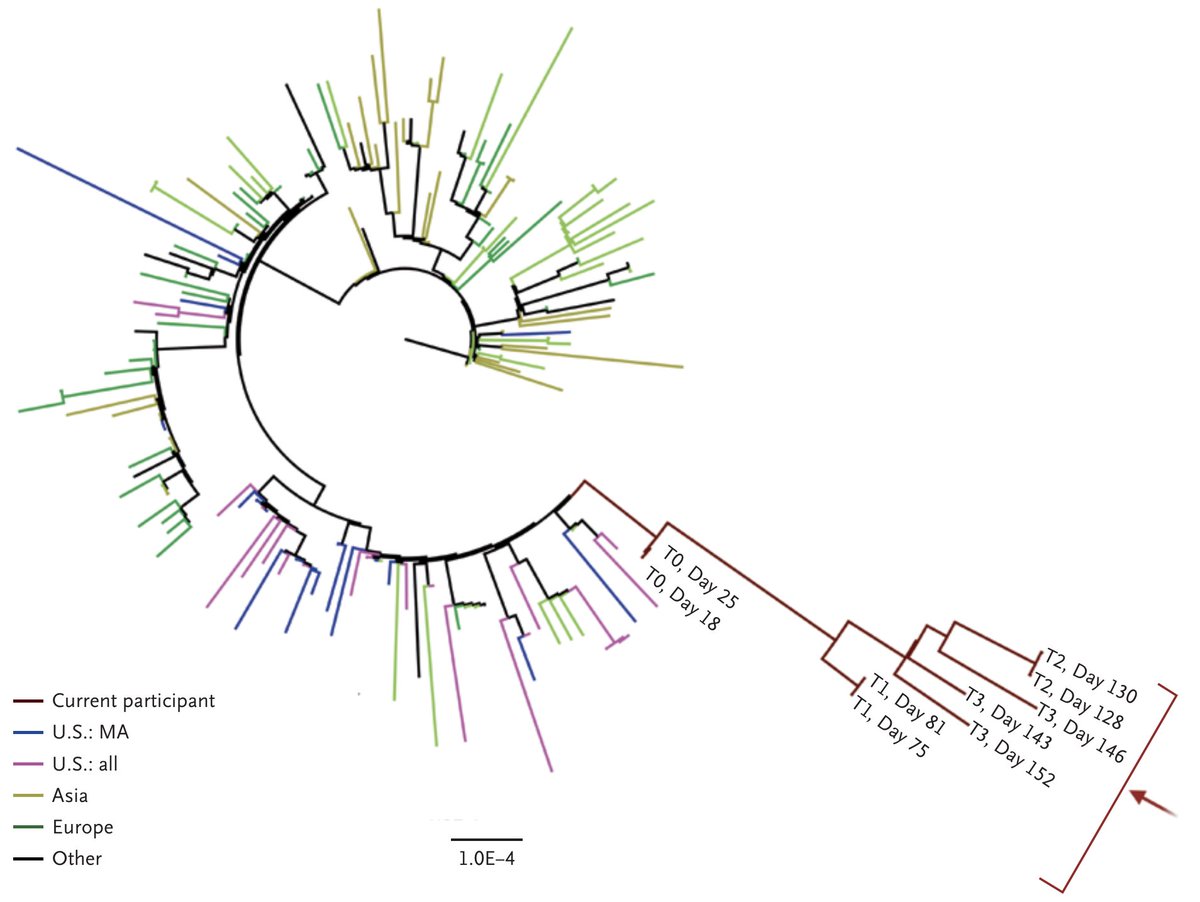
Indeed, this particular chronic infection evolved some of the exact same mutations seen in these variant viruses including 484K, 501Y and a 144 deletion. 16/19 
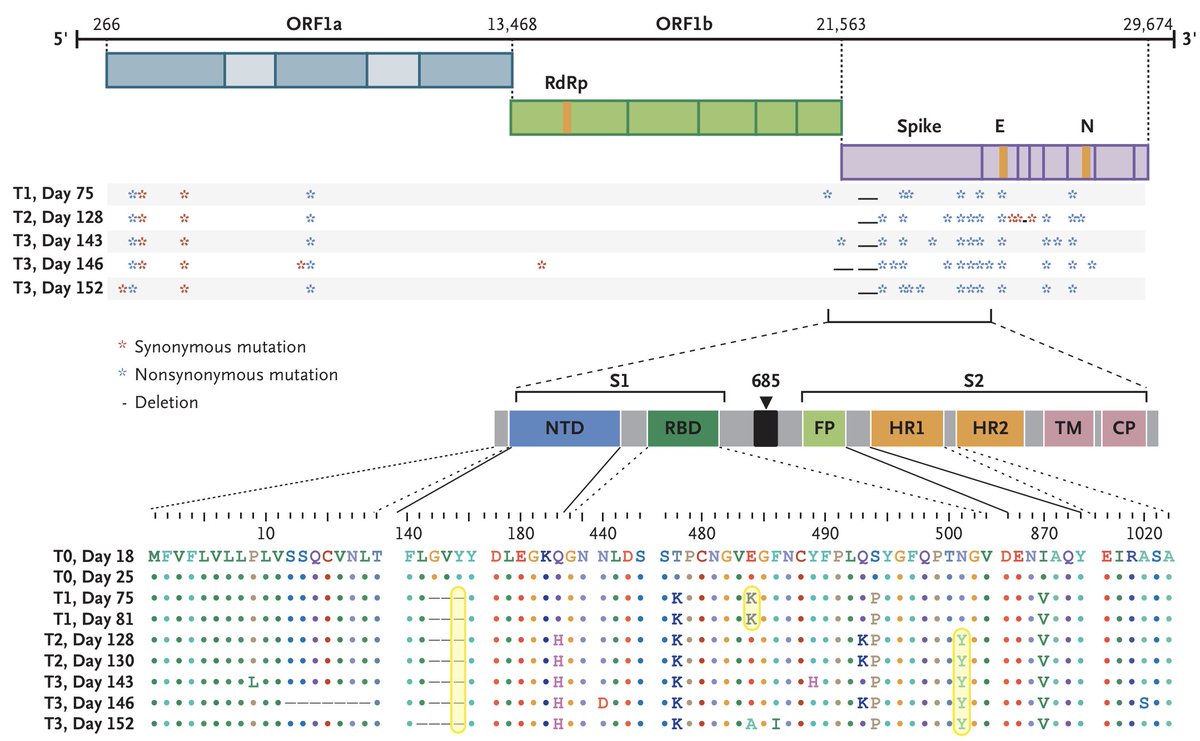
Additionally, @K_G_Andersen pointed me to work with coronavirus in mouse models showing that deletions are associated with chronic infections (ncbi.nlm.nih.gov/pmc/articles/P…). It's striking that the 3 AA deletion in ORF1ab is so far a hallmark of SARS-CoV-2 variant viruses. 17/19
Finally, the exceptionally long branch leading to 501Y.V1 is difficult to explain by a chain of infections given that genomic surveillance by @CovidGenomicsUK is sequencing approximately ~5% of cases; surveillance should have picked up intermediates if they existed. 18/19
Again, this hypothesis is highly speculative at this point, but separately, the fact that we've observed 3 variants of concern emerge since September suggests that there are likely more to come. 19/19
• • •
Missing some Tweet in this thread? You can try to
force a refresh