
Scientist @fredhutch, studying viruses, evolution and immunity. Collection of #COVID19 threads here: https://t.co/Yc4fun5rcp
246 subscribers
How to get URL link on X (Twitter) App

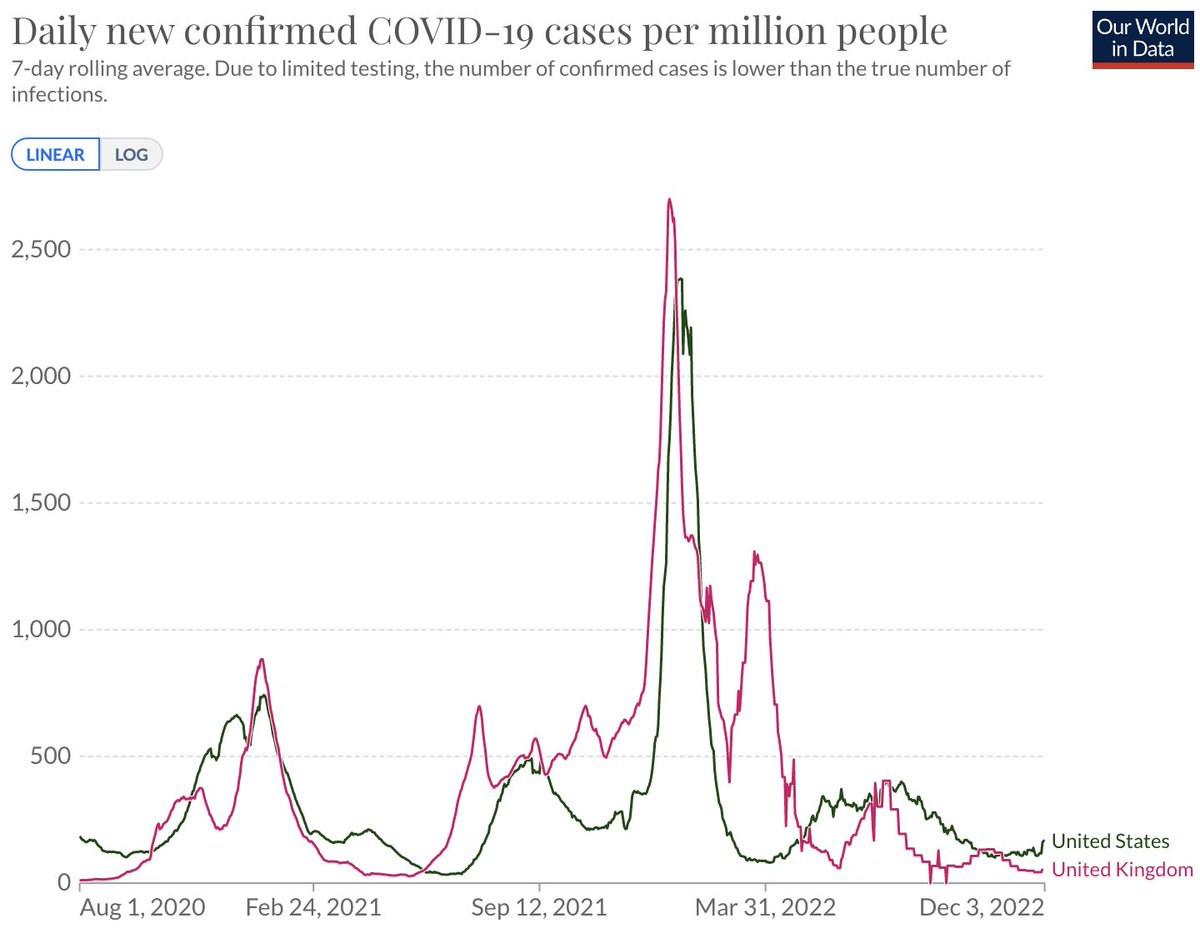
 However, at this point, nearly all infections will be in individuals with prior immunity from vaccination or infection and this combined with a roll back in testing makes it unclear how to interpret current case counts compared to previous time periods. 2/16
However, at this point, nearly all infections will be in individuals with prior immunity from vaccination or infection and this combined with a roll back in testing makes it unclear how to interpret current case counts compared to previous time periods. 2/16

 Lineage BA.2.75 (aka 'Centaurus') has been high on watch lists due to sustained increase in frequency in India combined with the presence of multiple mutations to spike protein. We now have enough sampled BA.2.75 viruses from outside India to make some initial conclusions. 2/10
Lineage BA.2.75 (aka 'Centaurus') has been high on watch lists due to sustained increase in frequency in India combined with the presence of multiple mutations to spike protein. We now have enough sampled BA.2.75 viruses from outside India to make some initial conclusions. 2/10

 It's necessarily fraught to try to make predictions of seasonal circulation patterns going forwards, but we can gain some intuition from simple epidemiological models. 2/11
It's necessarily fraught to try to make predictions of seasonal circulation patterns going forwards, but we can gain some intuition from simple epidemiological models. 2/11




 This is data from github.com/globaldothealt… and has had a 7-day smoothing applied and all y-axes are shown on a log scale. 2/10
This is data from github.com/globaldothealt… and has had a 7-day smoothing applied and all y-axes are shown on a log scale. 2/10



https://twitter.com/borges__vitor/status/1527436883093078029There are now over 300 confirmed and suspected cases globally and at least 15 viral genome sequences shared publicly. The extent and trajectory of the outbreak is still incredibly hazy. Here, I wanted to give a word of caution on genomic epidemiology of monkeypox. 2/9







 Looking at cases per 100k population per day across states, downturns are clear in NY, NJ, MA, FL, etc..., but many states are not yet at peak case loads. 2/9
Looking at cases per 100k population per day across states, downturns are clear in NY, NJ, MA, FL, etc..., but many states are not yet at peak case loads. 2/9 

 However, a large fraction of infections, symptomatic and otherwise, don't end up reported as cases due to lack of testing (either the individual doesn't seek testing or testing is desired but not readily found). 2/15
However, a large fraction of infections, symptomatic and otherwise, don't end up reported as cases due to lack of testing (either the individual doesn't seek testing or testing is desired but not readily found). 2/15

 We can partition state-level cases between Delta and Omicron using sequence data from @GISAID. This is made possible by 1.6 million genomes from the US from viruses sampled since Oct 15. This remarkable dataset is thanks to a large number of labs throughout the country. 2/12
We can partition state-level cases between Delta and Omicron using sequence data from @GISAID. This is made possible by 1.6 million genomes from the US from viruses sampled since Oct 15. This remarkable dataset is thanks to a large number of labs throughout the country. 2/12







https://twitter.com/trvrb/status/1467245887357210624Continuing previous methods, if we partition case counts from @OurWorldInData using sequence data from @GISAID and apply a modeling approach from @marlinfiggins we get rapid rises in Omicron cases in South Africa, Denmark, Germany, the UK and the US. 2/12



https://twitter.com/trvrb/status/1469157770716872708As before, we partition case counts from @OurWorldInData using sequences from @GISAID into estimated Omicron, Delta and other cases, and we use this partitioning to infer variant-specific Rt and epidemic growth rate r (methods and code here github.com/blab/rt-from-f…). 2/10

https://twitter.com/trvrb/status/1467245887357210624Because of significant travel connections (
https://twitter.com/MOUGK/status/1464141177226158094) and extremely strong genomic surveillance by @CovidGenomicsUK, we should have early data from the UK about rate of spread outside of South Africa. 2/21