With emerging variants of SARS-CoV-2 and initial evidence of antigenic evolution, I've seen comparisons here to seasonal influenza and its rate of evolution. In this thread, I want to ground these comparisons with some data. 1/18
https://twitter.com/trvrb/status/1340409968818671616
If we follow a transmission chain of SARS-CoV-2 from person to person, we'll generally see one mutation occur across the viral genome roughly every two weeks. 2/18
Here I use data from @nextstrain and @GISAID to compare sampling date to the number of mutations across the SARS-CoV-2 genome relative to initial genomes from Wuhan. This shows a steady accumulation of mutations through time with the average virus now bearing ~24 mutations. 3/18 
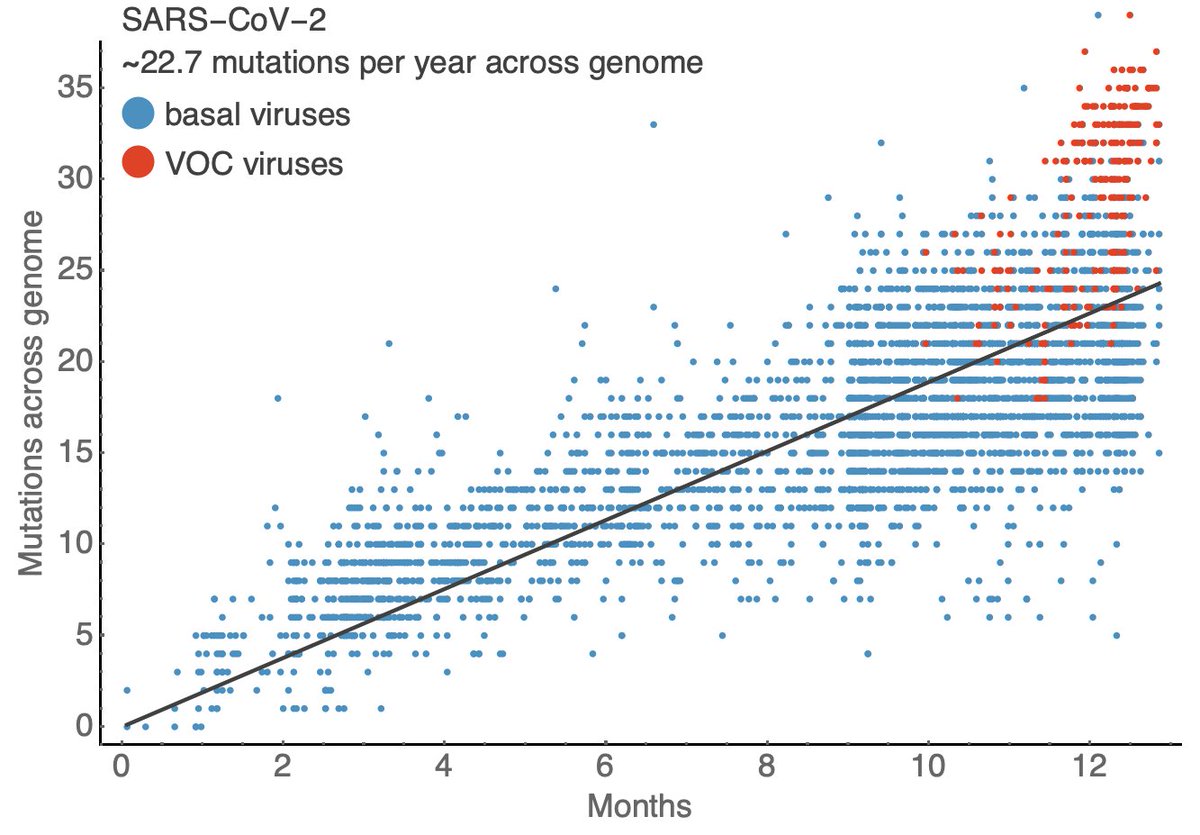
However, this is a random process and some viral lineages accumulate more mutations than others. In fact we see that the variants of concern (B.1.1.7 from the UK, 501Y.V2 from South Africa and P.1 from Brazil) possess more mutations than most circulating viruses. 4/18
The majority of mutations occurring in SARS-CoV-2 don't affect viral function and accumulate because the virus, as an RNA virus, undergoes error-prone copying. However, mutations that alter amino acids in the S1 portion of spike protein will often be functionally relevant. 5/18
This is a plot comparing sampling date to the number of amino acid changes in spike S1 subunit. Here we see a striking pattern where variant of concern (VOC) viruses show significantly more amino acid changes in spike S1 than other circulating viruses. 6/18 
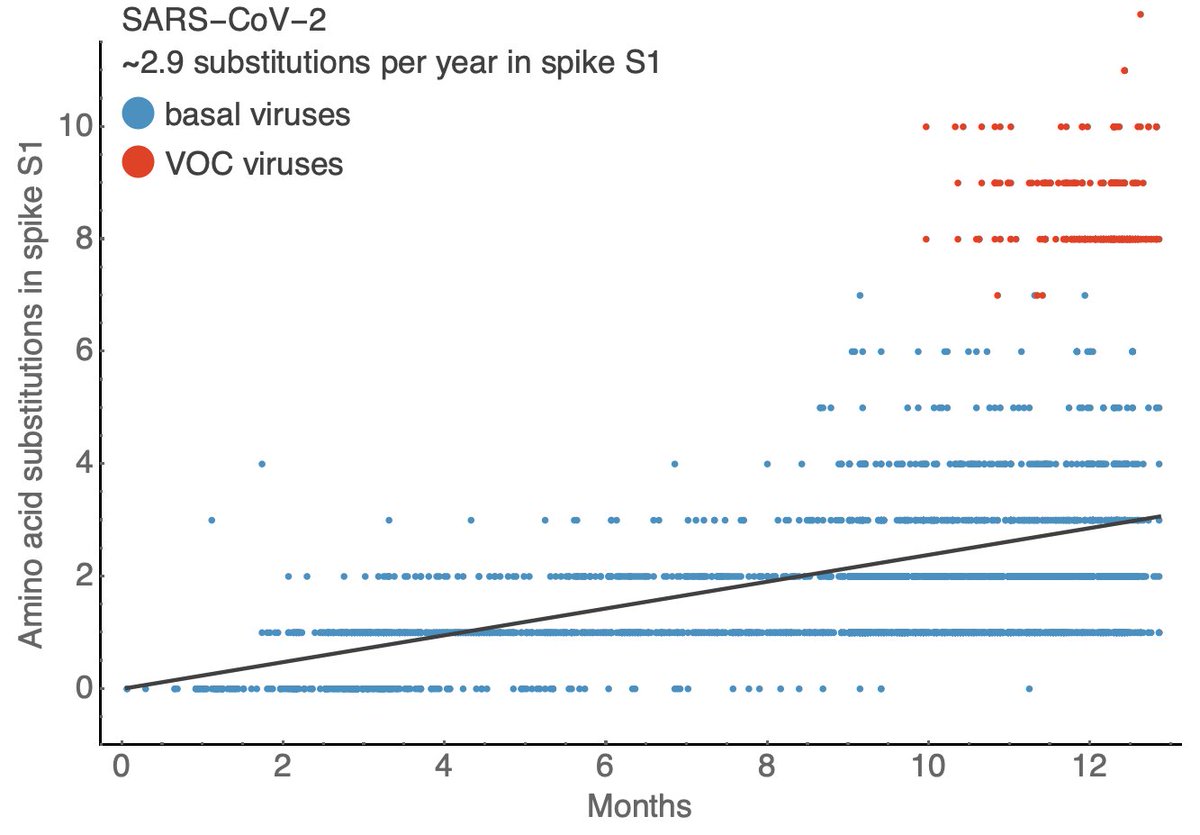
With VOCs included, on average SARS-CoV-2 is evolving at a rate of ~2.9 amino acid substitutions in spike S1 per year. This rate can be compared to the rate of amino acid changes in the HA1 domain of the surface protein HA in seasonal influenza. 7/18
There is a caveat in that these are different viruses and one amino acid change in spike S1 of SARS-CoV-2 may be more (or less) functionally relevant than one amino acid change in HA1 of seasonal influenza, but at least this gives a basis for a comparison. 8/18
And importantly the spike S1 subunit is 671 amino acids to HA1's 328 amino acids and so we might expect a ~2X rate difference just based on protein length as target for mutations. 9/18
For the fastest evolving seasonal influenza virus A/H3N2 we see a steady accumulation of ~2.5 amino acid substitutions per year over the past 12 years, which is slightly slower than what we're seeing now in SARS-CoV-2. 10/18 
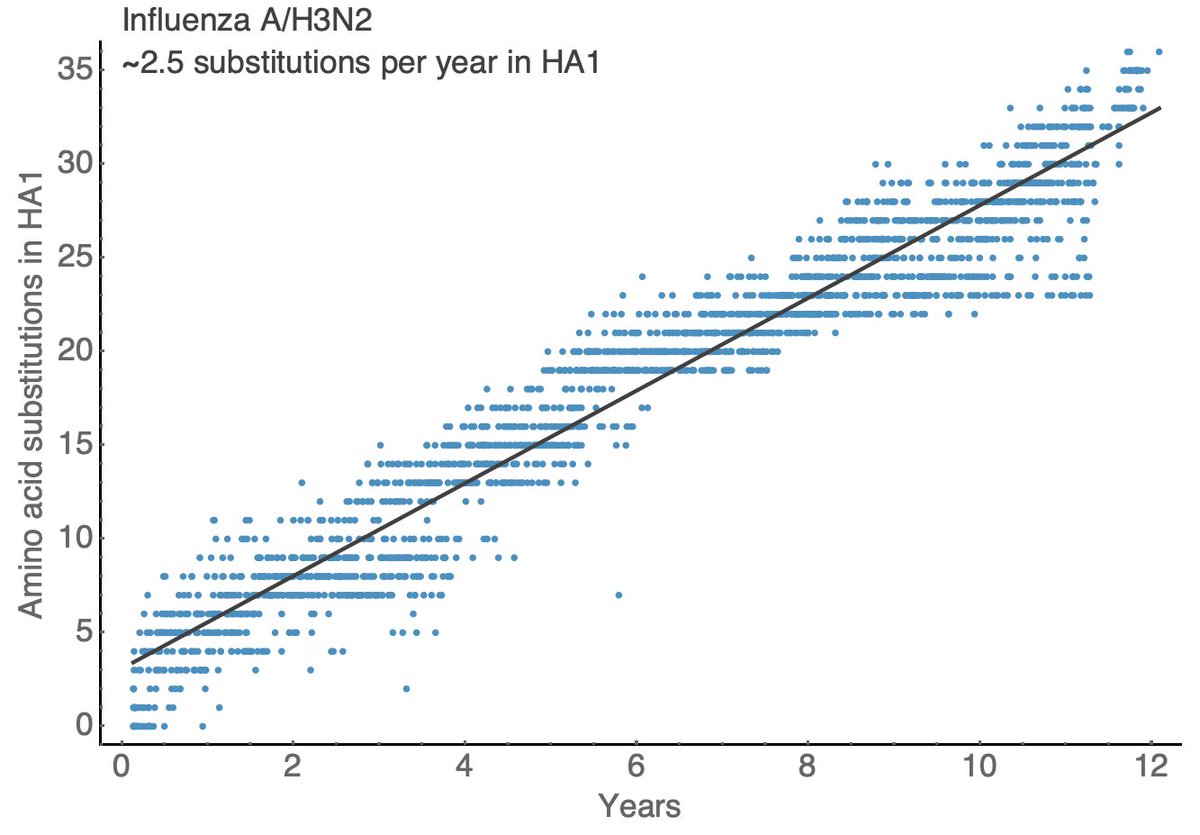
Other seasonal influenza viruses evolve more slowly with A/H1N1pdm showing ~1.5 substitutions per year in HA1, B/Vic showing ~0.5 substitutions per year in HA1 and B/Yam showing ~0.7 substitutions per year in HA1. 11/18 


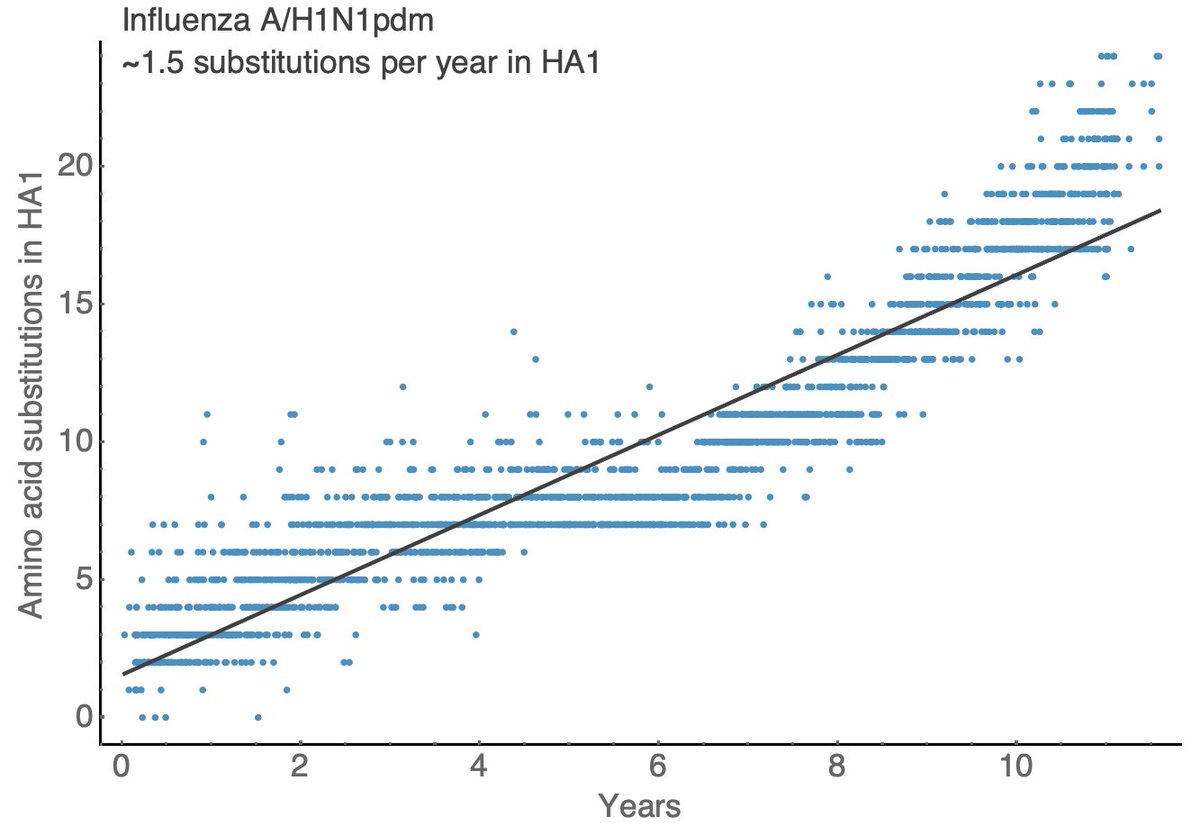
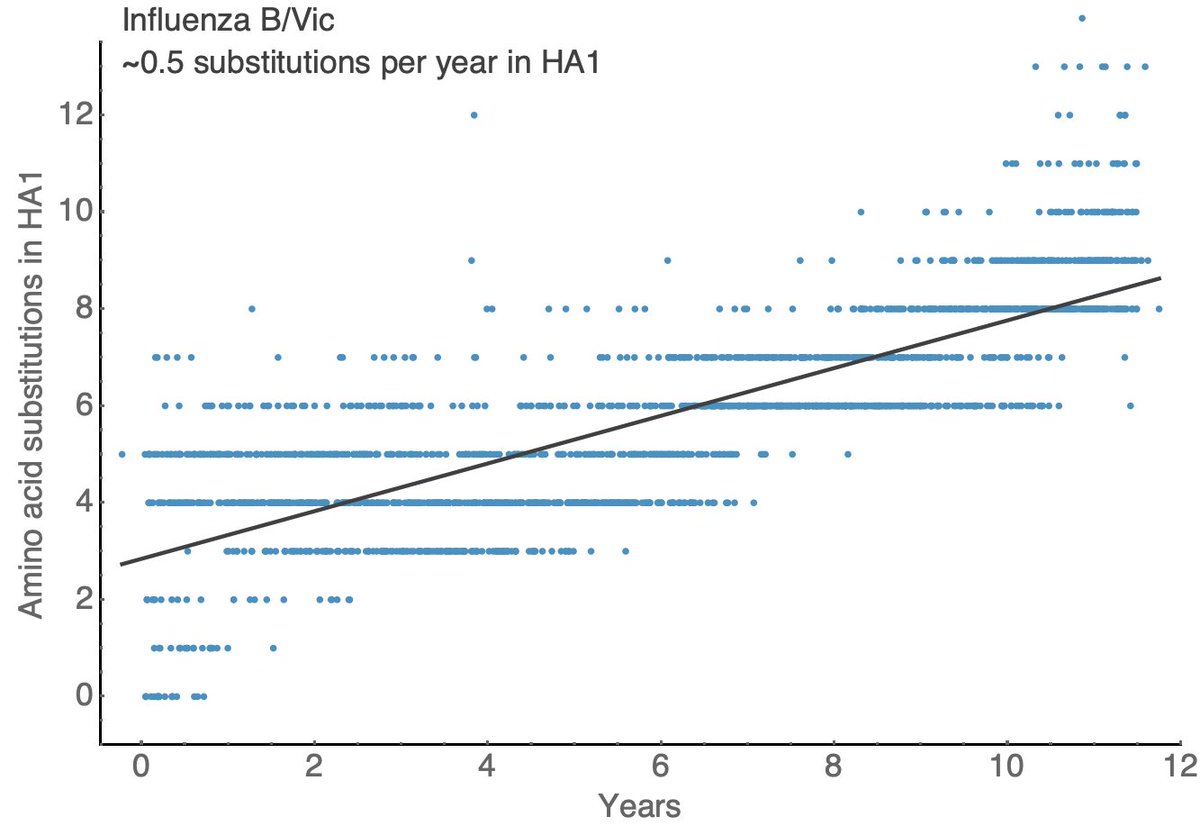
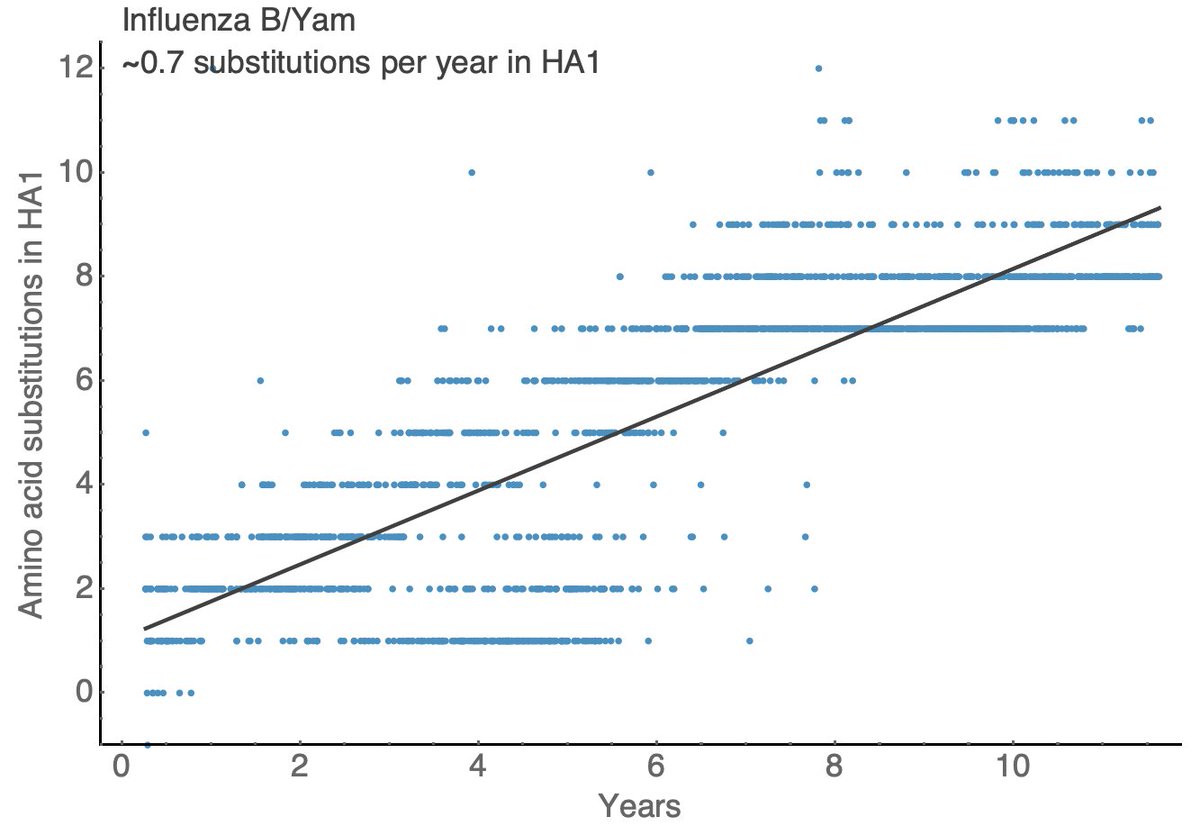
These rates of HA amino acid substitutions mirror experimentally determined rates of antigenic evolution. This is a figure from a 2014 paper by myself, @arambaut and others (bedford.io/papers/bedford…) showing faster antigenic drift in H3N2 than H1N1 than B/Vic and B/Yam. 12/18 
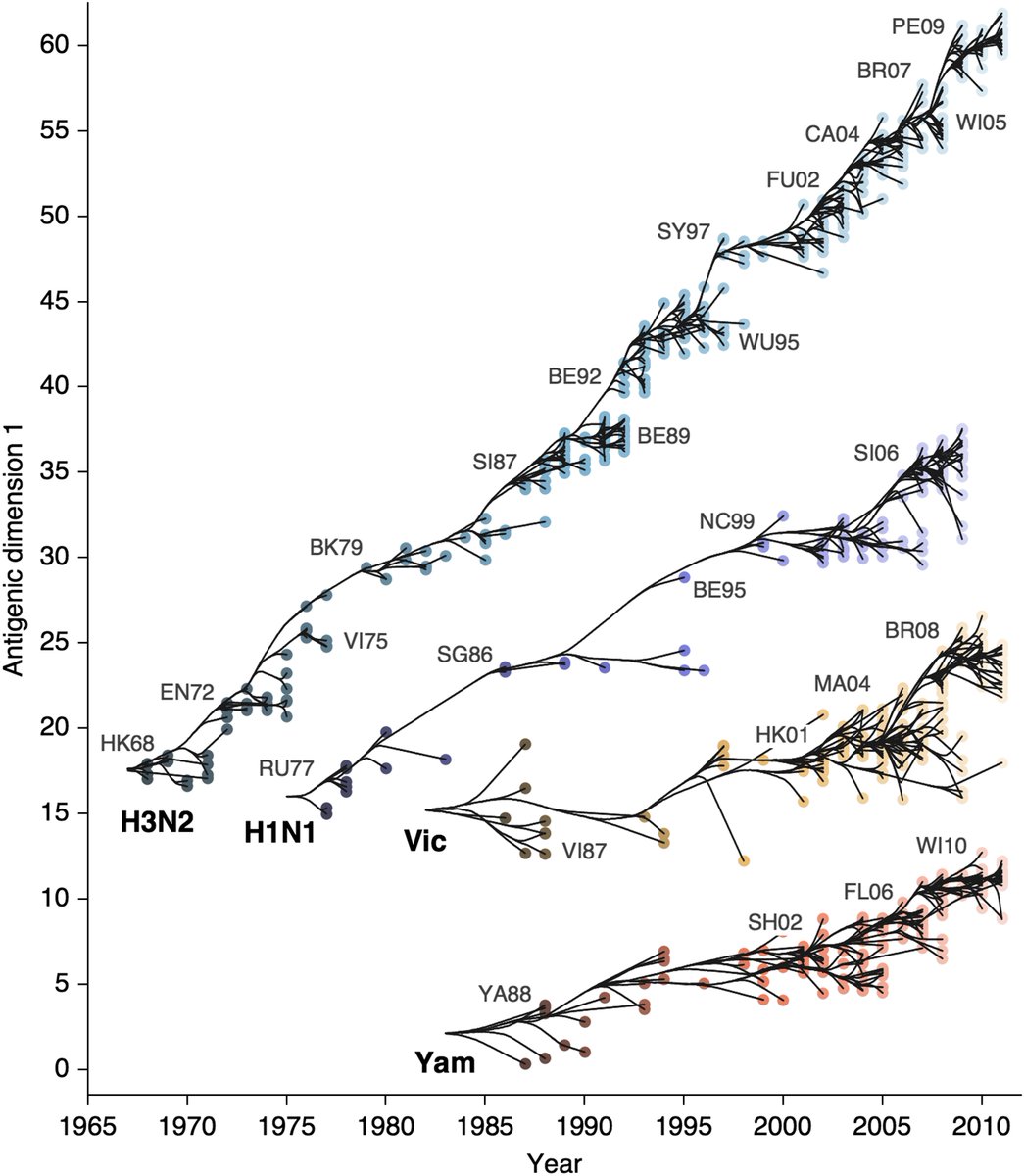
The rate of antigenic drift in influenza can be quantified by per-year fold-reduction in serological assays. For influenza H3N2, this rate averages ~1 two-fold titer reduction per-year. 13/18
The comparable datapoint for SARS-CoV-2 is work by Wibmer et al (biorxiv.org/content/10.110…) and Cele et al (medrxiv.org/content/10.110…) showing an ~8-fold titer reduction in neutralization assays to the 501Y.V2 variant from South Africa. 14/18
https://twitter.com/trvrb/status/1351785352793493505
This titer reduction is very roughly what is seen in an average of 3 years of influenza H3N2 evolution, but yearly jumps of 8-fold titer reductions in H3N2 are historically not uncommon, with H3N2 showing a staccato pace to its antigenic evolution. 15/18
Both simple rate of amino acid substitutions in spike S1 and titer drops in serological assays suggest that SARS-CoV-2 might be in the same ballpark as influenza A in terms of capacity for antigenic evolution. 16/18
That said, the evolution that we've seen with the recent variants of concern may represent an unusual circumstance in which the virus has made a large evolutionary jump to a new "fitness peak" and that won't be seen year-after-year. 17/18
Additionally, with new vaccine technologies (and particularly mRNA vaccines) we'll have the ability to more effectively chase the virus than we do with the seasonal influenza vaccine, which suffers from lower immunogenicity and longer lead times for strain updates. 18/18
Follow up #1: From March (
https://twitter.com/trvrb/status/1242628564324761606) until December, my expectation was antigenic evolution like in seasonal CoVs which are roughly as fast as flu B (see bedford.io/papers/kistler…). With VOCs, I now expect more like flu A, but this could be pace that's not sustained.
• • •
Missing some Tweet in this thread? You can try to
force a refresh