Evidence of rigged Peer Review @Eurosurveillanc
@PlanetWaves
@Bobby_Network
@RetractionWatch
@goddeketal
@PlanetWaves
@Bobby_Network
@RetractionWatch
@goddeketal
Admits to being the reviewer of the Corman-Drosten Retraction request.
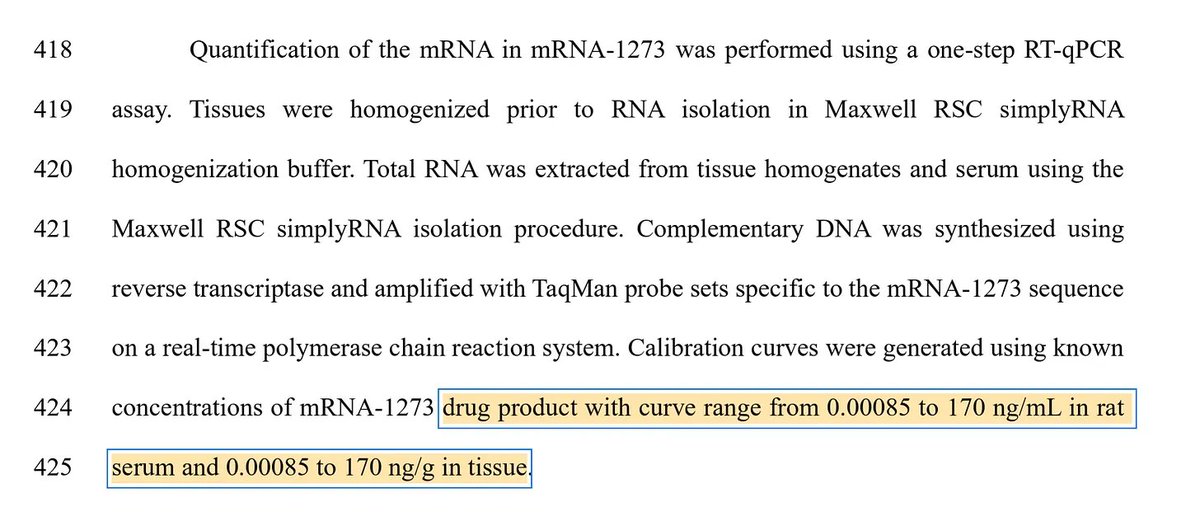
This is 60+ pages discussing Primer Dimers and other primer design problems.
But he just said he didn't look at the primers for this assay??
That is why Anon Peer Review is a Fraud.
@RetractionWatch
This is 60+ pages discussing Primer Dimers and other primer design problems.
But he just said he didn't look at the primers for this assay??
That is why Anon Peer Review is a Fraud.
@RetractionWatch
Bustin vs Andrew Wakefield.
Bustin testified under oath about the problems with Andrew Wakefields RT-qPCR. This is a big case.
It shares many of the same problems as Drosten?
OK for Drosten.
Not for Wakefield?
There is a difference.
Bustin was paid 220,000 pounds for it.
Bustin testified under oath about the problems with Andrew Wakefields RT-qPCR. This is a big case.
It shares many of the same problems as Drosten?
OK for Drosten.
Not for Wakefield?
There is a difference.
Bustin was paid 220,000 pounds for it.
What is it called when you contradict yourself multiple times under oath?
What is it called when you do it for 220,000 Pounds?
What is it called when you flip flop like this and it shuts down the world?
Still think Anon Peer Review is a good idea?
What is it called when you do it for 220,000 Pounds?
What is it called when you flip flop like this and it shuts down the world?
Still think Anon Peer Review is a good idea?
Bustin vs Wakefield
Bustin Vs Borger
Planet Waves Podcast
@PlanetWavesmega.nz/file/ocZAXBzS#…
cormandrostenreview.com
Bustin Vs Borger
Planet Waves Podcast
@PlanetWavesmega.nz/file/ocZAXBzS#…
cormandrostenreview.com
So was Wakefield rightfully scrutinized and Drosten given a hall pass?
Many of the concerns raised about Wakefields qPCR are significant. But the one most cited (contamination) by Dr. Bustin was not.
He claimed Wakefield omitted an RT-Step.
Bustin is emulating this in 2020.
Many of the concerns raised about Wakefields qPCR are significant. But the one most cited (contamination) by Dr. Bustin was not.
He claimed Wakefield omitted an RT-Step.
Bustin is emulating this in 2020.
Wakefield is also nailed for Lack of an SOP.
Same story in the Drosten review.
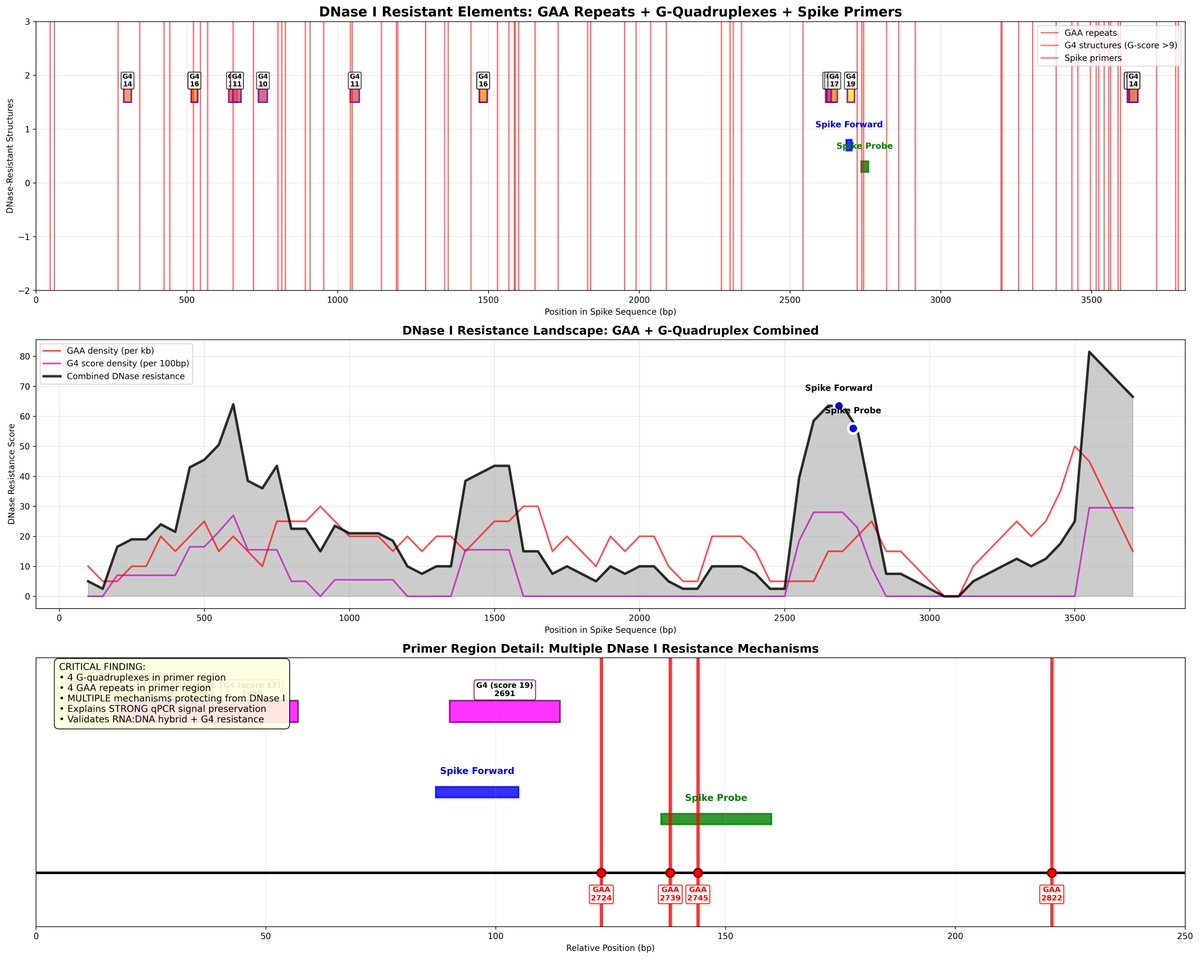
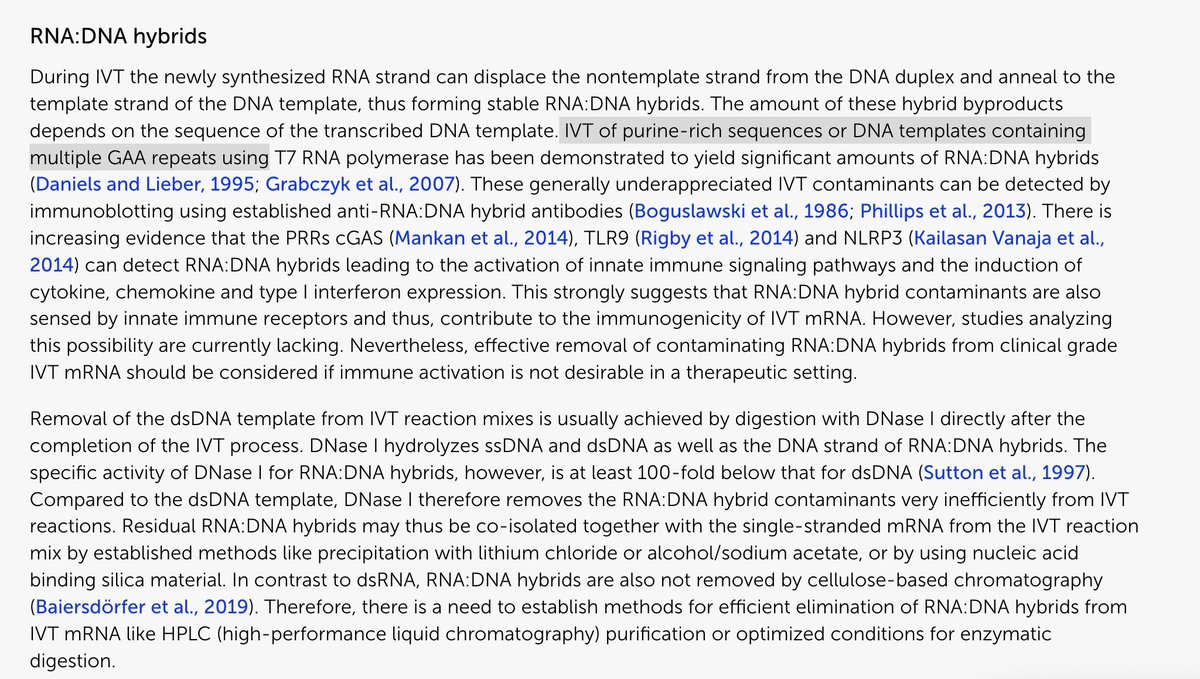
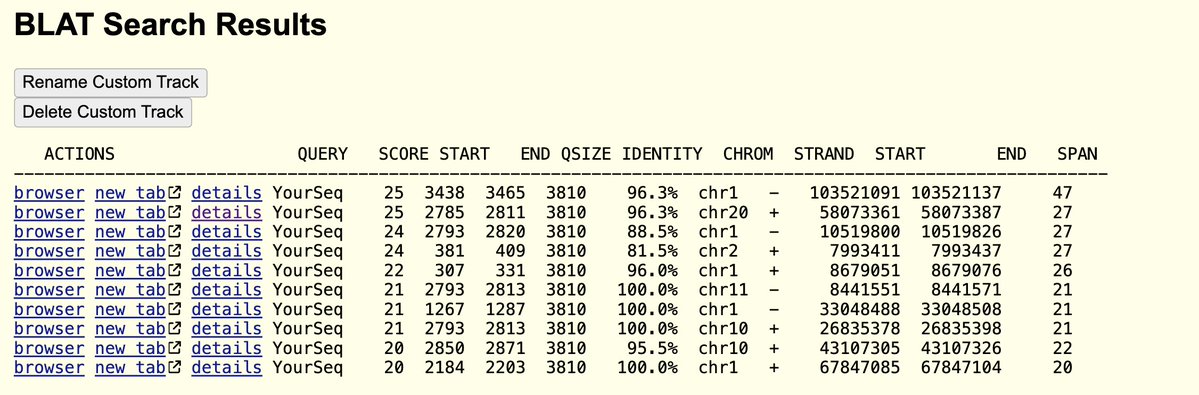
The proper way to address DNA or RNA contamination is with DNases and RNases.
RT enzymes are active at RTemp and as the cycler ramps its temperature up to 95C.
#BustinBusted
nature.com/articles/s4159…
Same story in the Drosten review.
The proper way to address DNA or RNA contamination is with DNases and RNases.
RT enzymes are active at RTemp and as the cycler ramps its temperature up to 95C.
#BustinBusted
nature.com/articles/s4159…
@threadreaderapp unroll
Bustin continues to push for limited to No RT step in qPCR.
Precisely what he persecuted Wakefield for…and was paid handsomely for the testimony he has since contradicted.


Precisely what he persecuted Wakefield for…and was paid handsomely for the testimony he has since contradicted.


@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh