
I gave our annual State of the Lab address this week! 🚀
I shamelessly borrowed the idea from Ken Dill--an incredibly clear communicator--who would annually dive into what we had done, how we got there, and where we were going.
Read on if you're interested in the gory details.
I shamelessly borrowed the idea from Ken Dill--an incredibly clear communicator--who would annually dive into what we had done, how we got there, and where we were going.
Read on if you're interested in the gory details.

What is our shared goal, as a lab?
We aim to transform drug discovery from a research enterprise into an engineering science.
Industrializing the production of new medicines is essential to enable us to have a significant positive impact on human health and disease.
We aim to transform drug discovery from a research enterprise into an engineering science.
Industrializing the production of new medicines is essential to enable us to have a significant positive impact on human health and disease.

It's well known that drug discovery has a high failure rate.
Circumstances may have improved a bit since this iconic depiction of the "failure cascade", but there's no debating that the overall success rate is low and costs are incredibly high.
nature.com/articles/nrd30…
Circumstances may have improved a bit since this iconic depiction of the "failure cascade", but there's no debating that the overall success rate is low and costs are incredibly high.
nature.com/articles/nrd30…

Drug discovery is unlike any other engineering discipline: We can design incredibly complex machines on computers, but not small molecule drugs.
Clearly, there's a disconnect between the predictive capability of models at the macroscopic scale and the microscopic scale.
Clearly, there's a disconnect between the predictive capability of models at the macroscopic scale and the microscopic scale.

There is a great deal we can learn from other disciplines that have made the transition from research (sometimes it works) to engineering (we can essentially guarantee it works). Bridge engineering is a fantastic example, with spectacularly high failure rates in the 1900s. 

Engineering failures are a fascinating subject, and I highly recommend this book to anyone who wants to learn more:
amazon.com/Design-Paradig…
amazon.com/Design-Paradig…
By taking the time to understand common modes of failure, we can make our models robust to them, and increase our success rates. This is tedious work, but without the discipline to do it, engineering fields could not exist. 

To succeed in our mission as a lab, we need to bring together some awesome people, do great science, disseminate it, and ultimately raise enough money to keep going. 

I am incredibly fortunate to have had the privilege of working with spectacular scientists. Several of them graduated in 2020. @andrrizzi was the first, moving to @groupparrinello where he could continue his stellar academic trajectory in his native Italy. 

Next to graduate was @SternChaya, one of the most driven scientists I have ever met. Chaya developed critically important methodologies that transformed the way @openforcefield builds force fields, and is now at @DeepCureAI helping discover new medicines. 

Third to graduate was @rafwiewiora, who taught us how to robustly construct Markov state models of protein dynamics, and is now a Redesign Science Distinguished Postdoctoral Fellow collaborating with @RedesignScience on COVID-19 drug discovery. 

Next, @_Mehtap_Isik_ carried out a tour de force in conceiving of, collecting data for, fielding, analyzing, reporting (as corresponding author), and even designing the journal covers for @samplchallenges that advanced our understanding of physical modeling for drug discovery. 

Now @_Mehtap_Isik_ is now a Computational Chemist at @moderna_tx, the same company that produced one of the first COVID-19 vaccines! 

Finally, Josh Fass (@maxentile) showed us how to understand uncertainty in molecular simulations, and taught us why MD actually works! He has now joined the fantastic scientists at @Relay_Tx working at the exciting interface of differentiable programming and drug discovery. 

For those of you curious why MD actually works, I highly encourage you to check out @maxentile's lucid article that makes complex concepts---such as Langevin integrators and timestep-dependent errors---highly comprehensible!
choderalab.org/publications/2…
choderalab.org/publications/2…

The most important part of my job (and the best part!) is making sure everyone in the lab goes on to a fulfilling career that makes use of their unique skills. I couldn't be happier with how folks who moved on in 2020 went on to do amazing things. 

We've got a fantastic crew aboard right now, even though we're more distributed due to COVID. On the other hand, this means we've been able to tap incredibly talented people we wouldn't have been able to otherwise.
You can read about all of them at:
choderalab.org/members
You can read about all of them at:
choderalab.org/members

We're also incredibly fortunate to be collaborating with the rising superstar Andrea Volkamer at the @ChariteBerlin, funded by the @Einstein_Berlin and @berlinnovation, which supports research and travel exchanges between the groups.
volkamerlab.org
volkamerlab.org

We are humbled to be part of the @openforcefield Initiative, led by @karmecon @davidlmobley @MichaelKGilson @Michael_Shirts. This wonderful group of researchers and software scientists is a template for how we can solve major challenges in our field.
openforcefield.org
openforcefield.org

We have so many other incredible collaborators that it's hard to name (or tag!) them all! Suffice it to say we're extremely grateful to work with so many talented scientists in academia and industry who are supportive of open science and open source software to advance the field.
Seven years in, we're still not perfect. But we're dedicated to consistently improving everything about the lab and how it runs. We subscribe to the principle of kaizen ("continual improvement"), and have a dedicated slack channel to discussing how we can always aim to do better. 

We currently use a variety of coordination tools to do our work, including @googlenonprofit, @github, @SlackHQ, @NotionHQ, @overleaf, @zotero, @trello, and @OpenEyeSoftware. We couldn't do it without these tools! 

We periodically remind everyone of these policies (and now have them in our lab handbook on @NotionHQ), but nothing has drilled in just how important it is to have flexible work policies as much as COVID has! 

Thanks to our awesome lab manager Erica, we have a new awesome @NotionHQ workspace that helps us organize our COVID research and all other lab operations! 

Our lab builds and contributes to a number of open source tools that aim to make physical modeling accessible and useful in drug discovery or biomolecular research. We're thrilled to contribute to @openmm_toolkit and its ecosystem of tools.
choderalab.org/code
choderalab.org/code

In addition to foundation libraries and toolkits, we build applications tailored to exploring or solving challenges in specific areas. Often these codes are rather experimental, but some go on to be production codes! 

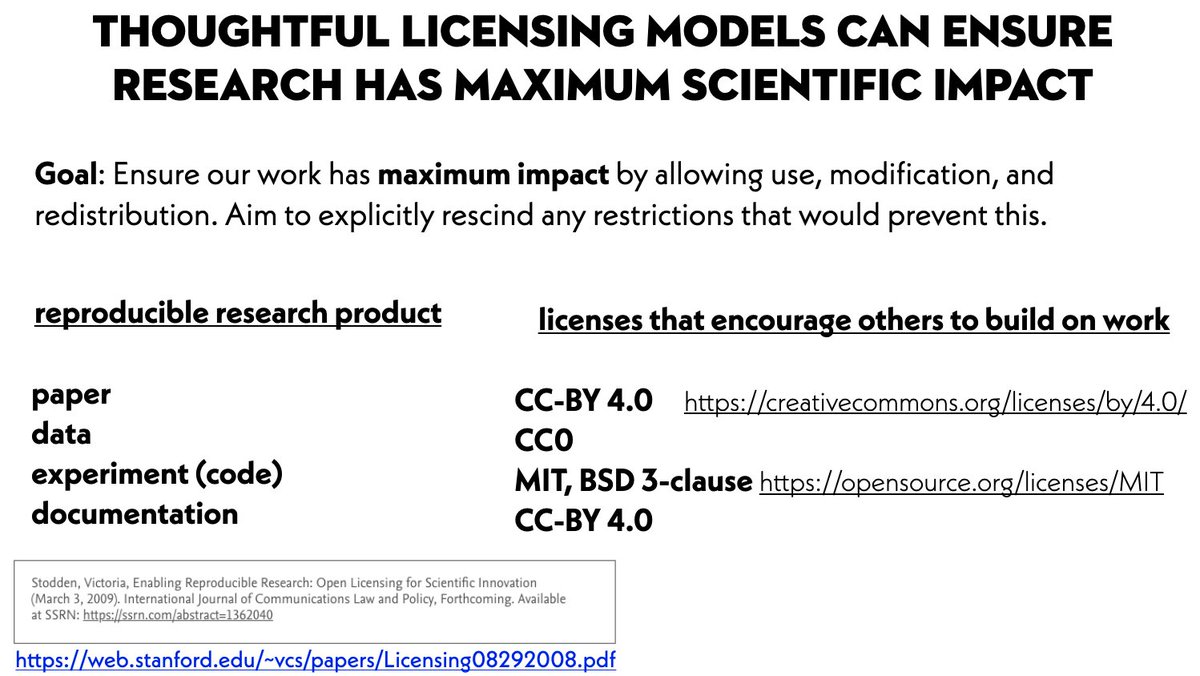
We aim for our work to have maximum impact by ensuring everyone is free to use, modify, build on, and distribute our work. Thankfully, the brilliant @victoriastodden has established best practices for reproducible research:
web.stanford.edu/~vcs/papers/Li…
web.stanford.edu/~vcs/papers/Li…
As @wpwalters consistently points out, other fields celebrate making research reproducible by releasing code with papers. We believe our field should do the same as part of the reproducible research product.
paperswithcode.com
paperswithcode.com

We are heavily inspired by how productive machine learning has become once enabled by well-supported, high-performance toolkits with powerful, extensible, composable abstractions. We aim to follow these principles in designing our own tools. 

openmmtools, a batteries-included library for @openmm_toolkit mostly written by the fantastic @andrrizzi, is an attempt at creating powerful abstractions. This is an entire replica-exchange binding free energy code with enhanced sampling! openmmtools.readthedocs.io/en/0.18.1/gett… 

We are working to codify our principles and practices online, and harmonize them across the community (with trailblazing organizations like @openforcefield) to break down barriers to collaboration within our field.
github.com/choderalab/sof…
github.com/choderalab/sof…

For those not familiar with how useful software abstractions can be, I liken this to the "Taco Bell" model of software development: Different combinations of the same ingredients can produce many different resulting products! 

For those looking for more useful abstractions on top of OpenMM that can easily be combined in new ways to develop powerful applications, definitely check out openmmtools!
openmmtools.readthedocs.io
openmmtools.readthedocs.io

Much of my 2020 was occupied by working with the amazing scientists that have come together for the @covid_moonshot, which aims to develop a new oral small molecule therapy for COVID-19. You can check out how far we've come in my recent talk: choderalab.org/news/covid-moo… 

There's so much amazing science going on that I can't possibly pack it into a tweet thread, so I'll skip to the highlights of how we've disseminated some of what we did in 2020. 

We're huge believers in preprints, and try to preprint everything we do---often far ahead of submission, to solicit feedback from the community---as well as post it on our group website:
choderalab.org/publications/
choderalab.org/publications/

I'm honestly not sure how this happened, and it must have mostly been driven by all the fantastic scientists in the lab rather than me (since I was spending most of my time trying to put out fires), but we published or preprinted 14 papers in 2020.
choderalab.org/publications
choderalab.org/publications
1/ @rafwiewiora, along with @cwhmyr @FrankNoeBerlin, and Ernesto Suarez, dove deep into what Markov state models can and cannot tell us about biomolecular dynamics:
choderalab.org/publications/2…
choderalab.org/publications/2…

2/ @wiedermc and @maxentile showed us that not only can we carry out alchemical free energy calculations with quantum machine learning potentials, we can train them to both QM and experimental data to significantly improve prediction from minimal data!
choderalab.org/publications/2…
choderalab.org/publications/2…

3/ @YuanqingWang and @maxentile also showed us how to rethink the entire way MM force field parameters are applied, demonstrating we can use graph convolutional networks to make the whole process end-to-end differentiable! This is totally transformative.
choderalab.org/publications/2…
choderalab.org/publications/2…

4/ @SternChaya, working with fantastic scientists from @openforcefield (including @maxhostile @dga_smith @davidlmobley), showed us how to fragment molecules to preserve chemical environments for quantum chemical calculations in building force fields.
choderalab.org/publications/2…
choderalab.org/publications/2…

5/ I was thrilled to be part of an all-star group led by @ppxasjsm (with @hannahbruce @julienmich80 @davidlmobley @LeviXansNaden @andrrizzi @Jenkescheen @Michael_Shirts @gtresadern @hardsphere) on best practices for free energy calculations for @livecomsj
choderalab.org/publications/2…
choderalab.org/publications/2…

6/ In work that totally blew my mind, Dom Rufa and @hannahbruce led work (with @adrian_roitberg @olexandr) that demonstrated replacing ligand intramolecular energetics with ANI2x can cut the error in ligand binding free energy calculations in half! 🤯
choderalab.org/publications/2…
choderalab.org/publications/2…

7/ Early in the pandemic, @iamalphalee, @london_lab, @FrankvonDelft and I penned a call for scientists to join our crowdsourced effort to develop a new small molecule oral inhibitor for COVID-19 in @NatureChemistry, and the response was overwhelming!
choderalab.org/publications/2…


choderalab.org/publications/2…



8/ @steven_albanese's @Schrodinger internship paper on how free energy calculations can be *more* accurate for selectivity prediction than affinity prediction due to correlations in force field errors finally came out!
choderalab.org/publications/2…
choderalab.org/publications/2…

9/ @foldingathome became the first computing platform to cross the exaFLOP/s barrier as amazing humans around the world supported our efforts to aid experimental colleagues in developing new COVID-19 therapies, documented in this @drGregBowman preprint.
choderalab.org/publications/2…
choderalab.org/publications/2…

10/ @Ana_J_Silveira and @rafwiewiora contributed to an awesome survey of how machine learning was transforming molecular simulation.
choderalab.org/publications/2…
choderalab.org/publications/2…

11/ After fielding our first blind pKa prediction @samplchallenges, our awesome collaborator Marilyn Gunner came to the realization there was a brilliantly simple way to summarize the essential information for reporting small molecule pKas.
choderalab.org/publications/2…
choderalab.org/publications/2…

12/ @_Mehtap_Isik_ reported the results and lessons learned from the first octanol-water logP @samplchallenges, with contributions from many others!
choderalab.org/publications/2…
choderalab.org/publications/2…

13/ @andrrizzi did a phenomenal job in fielding and assessing the first @samplchallenges SAMPLing challenge, meant to assess how rapidly different codes converged to the true force field free energy, but instead finding that no two codes could agree!
choderalab.org/publications/2…
choderalab.org/publications/2…

14/ And finally, @_Mehtap_Isik_ and fantastic collaborators from @Merck Rahway (and @davidlmobley) wrote a fantastic paper on logP measurements for a #compchem audience in describing how data was collected for @samplchallenges.
choderalab.org/publications/2…
choderalab.org/publications/2…

Preprints really do work. They allow us to share our work rapidly, and eliminate barriers to access. From the number of downloads, they seem to be incredibly effective tools for driving dissemination.
biorxiv.org/search/author1…
biorxiv.org/search/author1…

• • •
Missing some Tweet in this thread? You can try to
force a refresh







