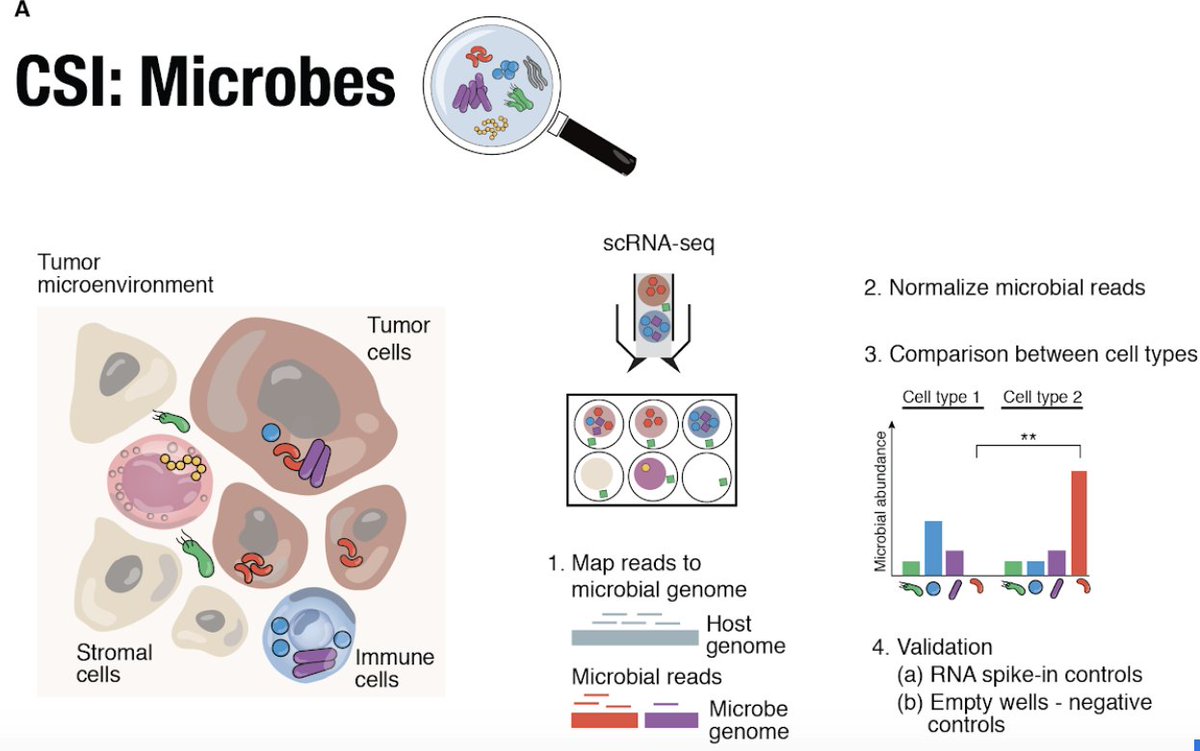
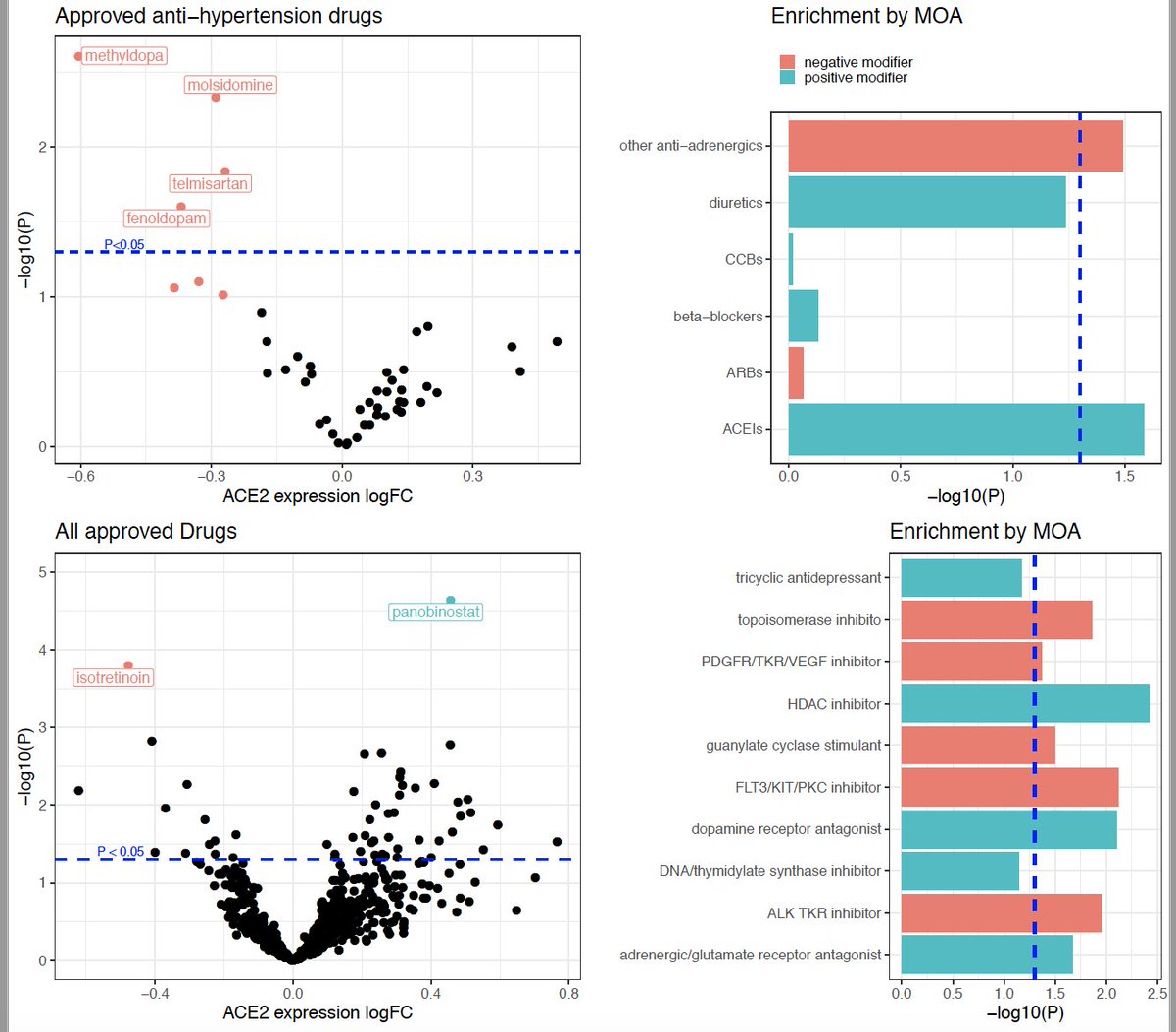
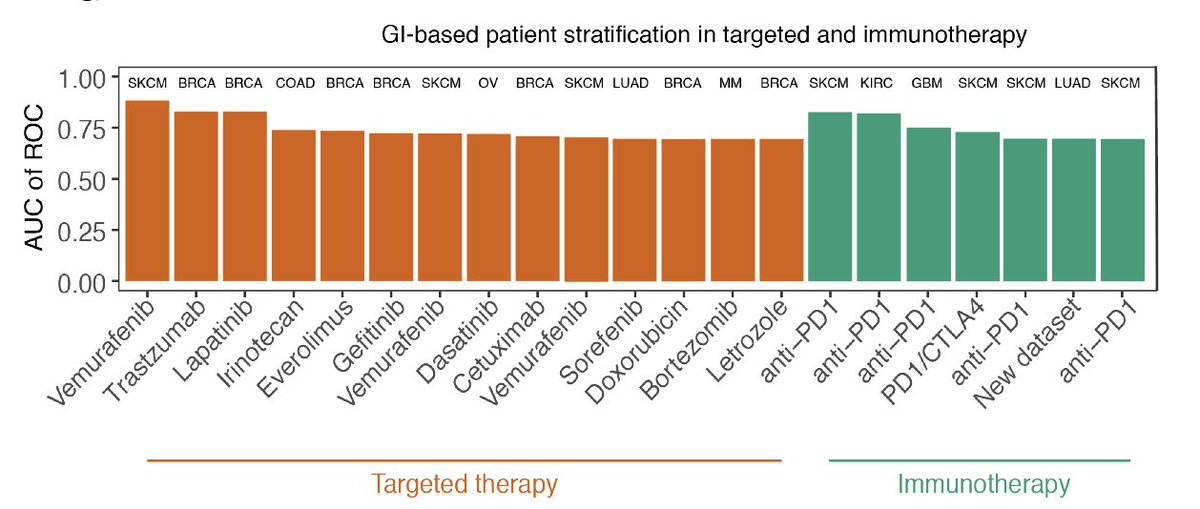
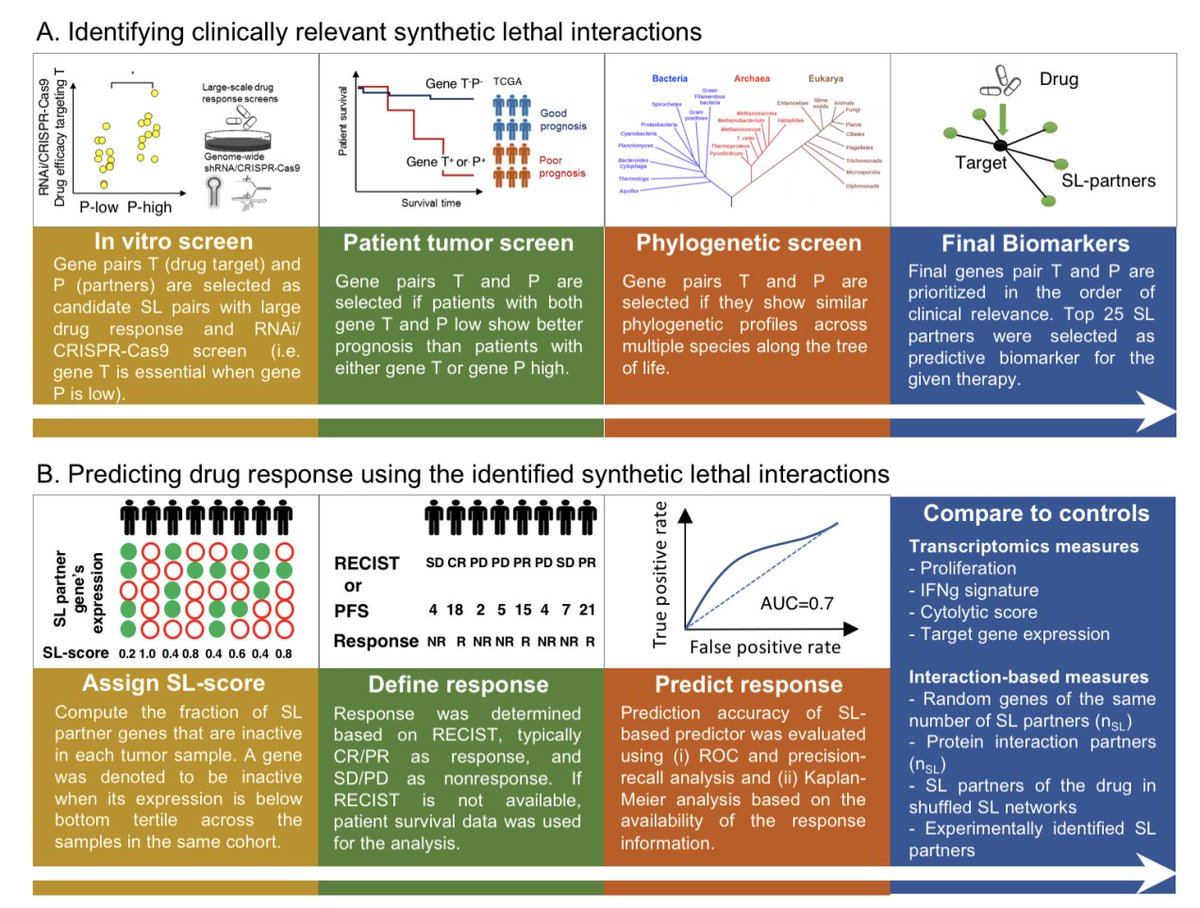
Can we infer an individual’s tissue-specific expression from their whole blood transcriptome?
Hot off the Press & a summary of our effort is below!
advances.sciencemag.org/content/7/14/e…
with @hannenhalli & @KunWang331
Hot off the Press & a summary of our effort is below!
advances.sciencemag.org/content/7/14/e…
with @hannenhalli & @KunWang331

Learning an individual’s tissue-specific gene expression can be invaluable guiding diagnosis & monitoring progression of a wide range of diseases. Unfortunately, for most tissues, the transcriptome cannot be obtained without invasive procedures.
In contrast, measuring whole blood expression is minimally invasive and high time resolution.
Thus, we asked a natural but quite challenging question - whether and to what extent can we predict an individual’s tissue-expression (for 32 tissues) based solely on their whole blood gene expression.
Approach: Using Genotype-Tissue Expression (GTEx) database with expression profile of a wide-range of tissues and matched whole blood, we built a generalized linear model for each gene expression in tissue from Whole blood-expression.
We were significantly able to (P<0.05) predict tissue-specific expression levels for ~60% of the genes on average across 32 tissues. 

In a downstream analysis, we found the tissue-specific expression inferred from the blood transcriptome is almost as good as the actual measured tissue expression in predicting disease state for six different complex disorders. 

In sum, we present a computational approach and proof-of-concept for a pipeline to predict tissue-specific expression using whole blood expression. Will leave the rest for your read.
• • •
Missing some Tweet in this thread? You can try to
force a refresh