We are excited to announce our paper in @NatureGenet uncovering a role for minor introns in clonal hematopoietic disorders, Noonan Syndrome, and a diverse number of cancers. A collaboration between our lab (@sloan_kettering) and @bradleybio (@fredhutch): nature.com/articles/s4158…
2.Most eukaryotes have 2 splicing machineries: the major & minor spliceosome. The minor spliceosome recognizes <0.5% of the introns in the human genome. Since their discovery, biological roles for minor introns have been enigmatic. 
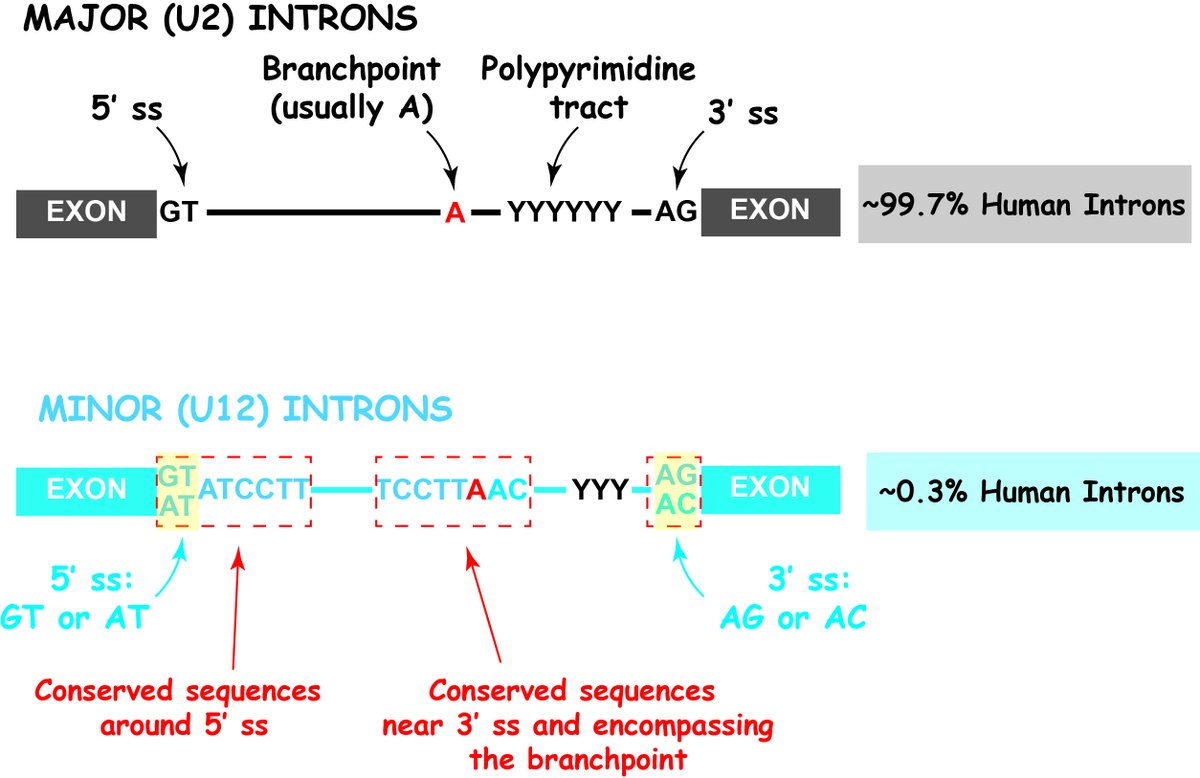
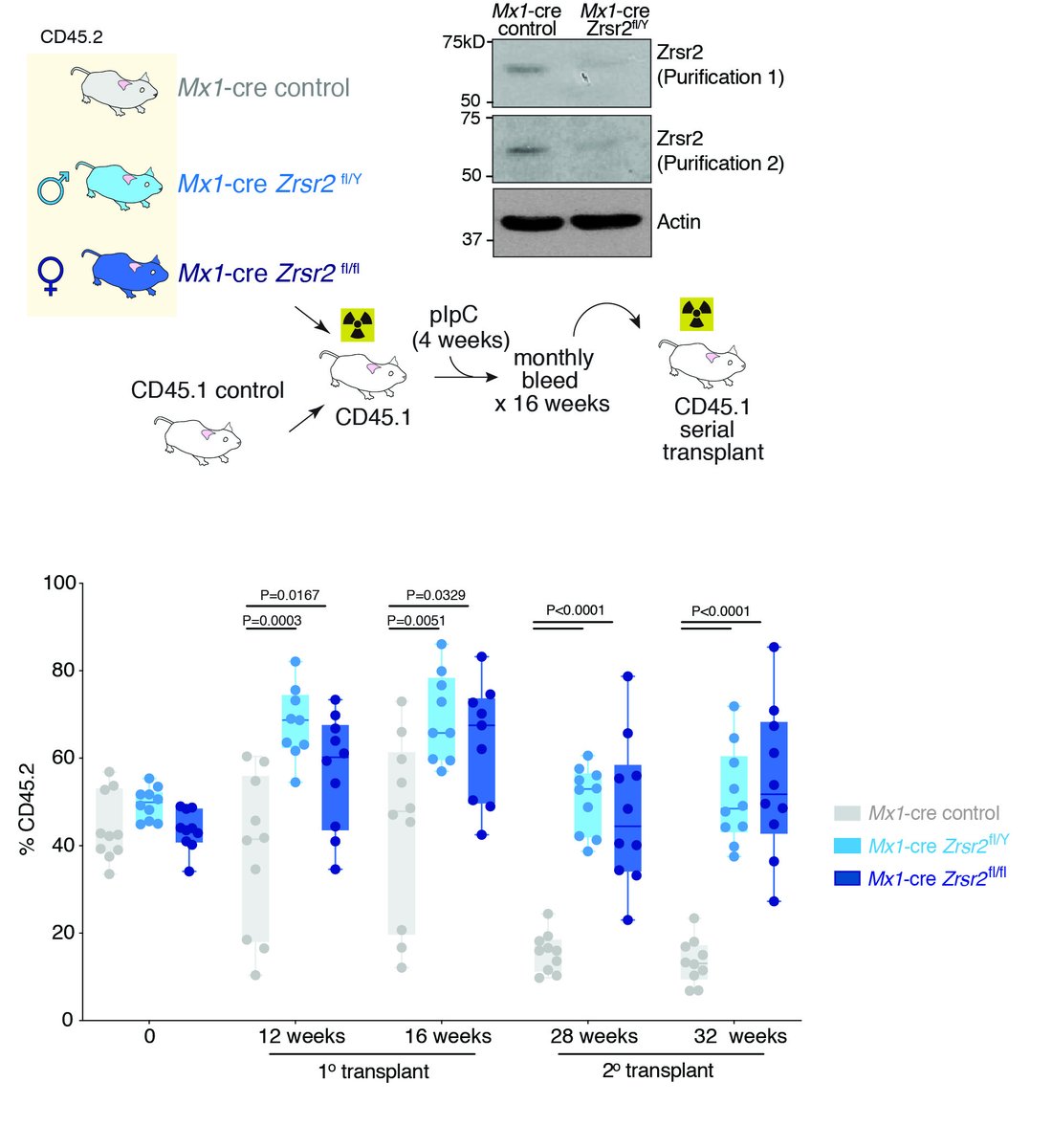
3.In work co-led by @DaichiInoue5, Jacob Polaski, and @TaylorJ_MD we identify that deletion of the minor spliceosome regulatory protein ZRSR2, frequently mutated in MDS and AML, enhances hematopoietic stem cell self-renewal. 
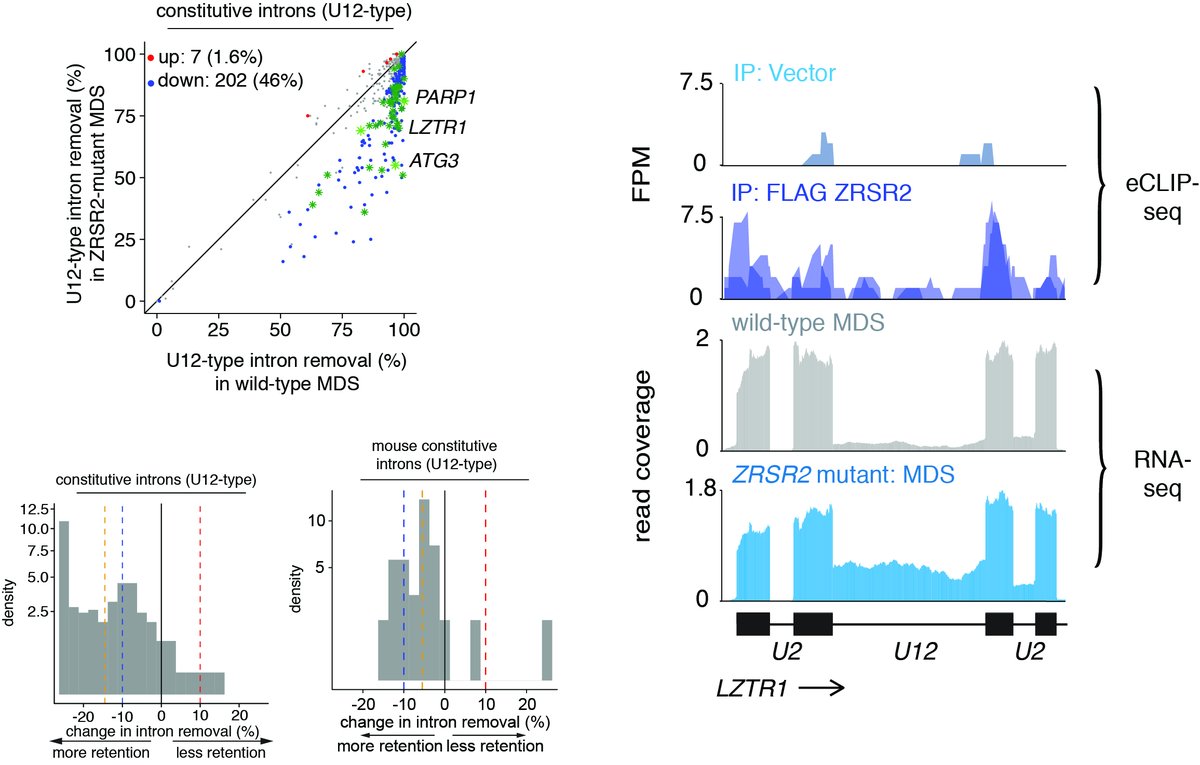
4.We mapped ZRSR2 responsive introns across patients and mouse hematopoietic cells using RNA-seq and anti-ZRSR2 eCLIP-seq to find a subset of ZRSR2 regulated introns. 
5.Beautiful work from Jose Pineda @bradleybio identified unique branchpoint features of ZRSR2 regulated introns. Please see Jose’s prior great paper in @GenesDev:
genesdev.cshlp.org/content/early/…
genesdev.cshlp.org/content/early/…
6.We then performed a CRISPR functional enrichment screen to identify those ZRSR2 regulated splicing events which may be associated with clonal advantage in hematopoietic cells. 
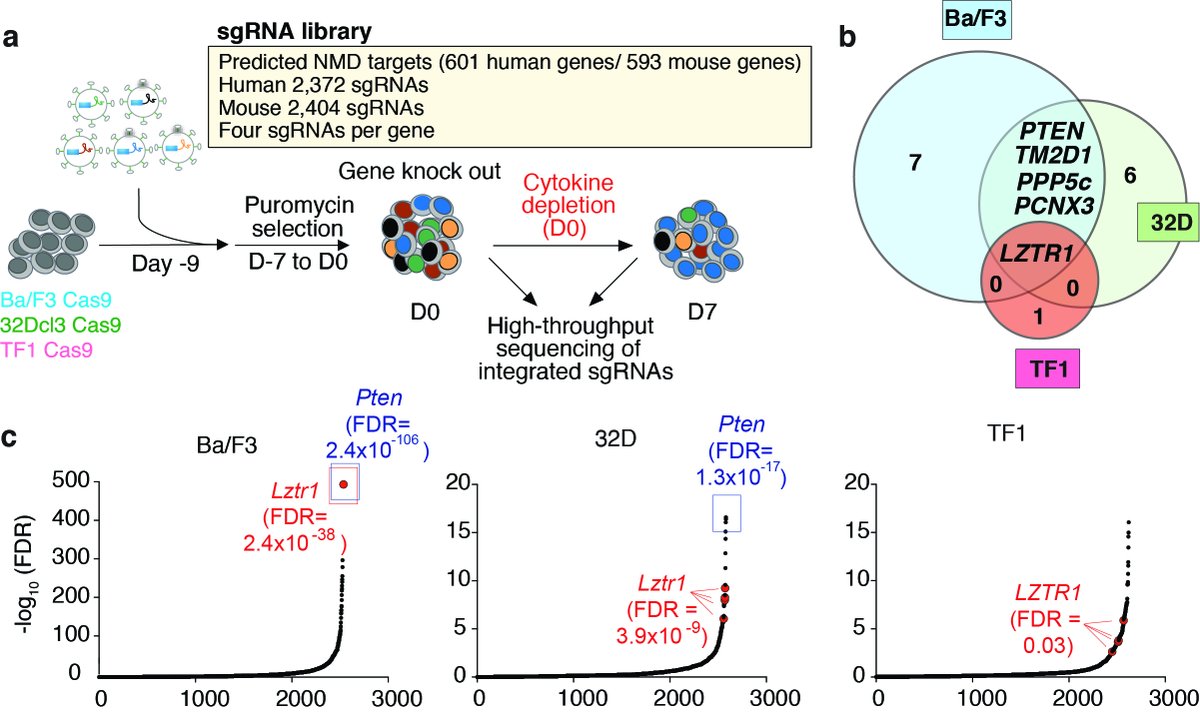
7.This revealed a surprising discovery of mis-splicing of the minor intron of the CUL3 adaptor regulating RAS GTPas abundance LZTR1 in ZRSR2-mutant MDS and AML.
8.With wonderful help from @castel_pau & the Frank McCormick lab (@frankpmccormick) we found that mutations within LZTR1’s minor intron itself was transforming to hematopoietic cells and present in Noonan Syndrome (@noonansyndrome, @Noonan_Syndrome) as well as Schwannomatosis. 
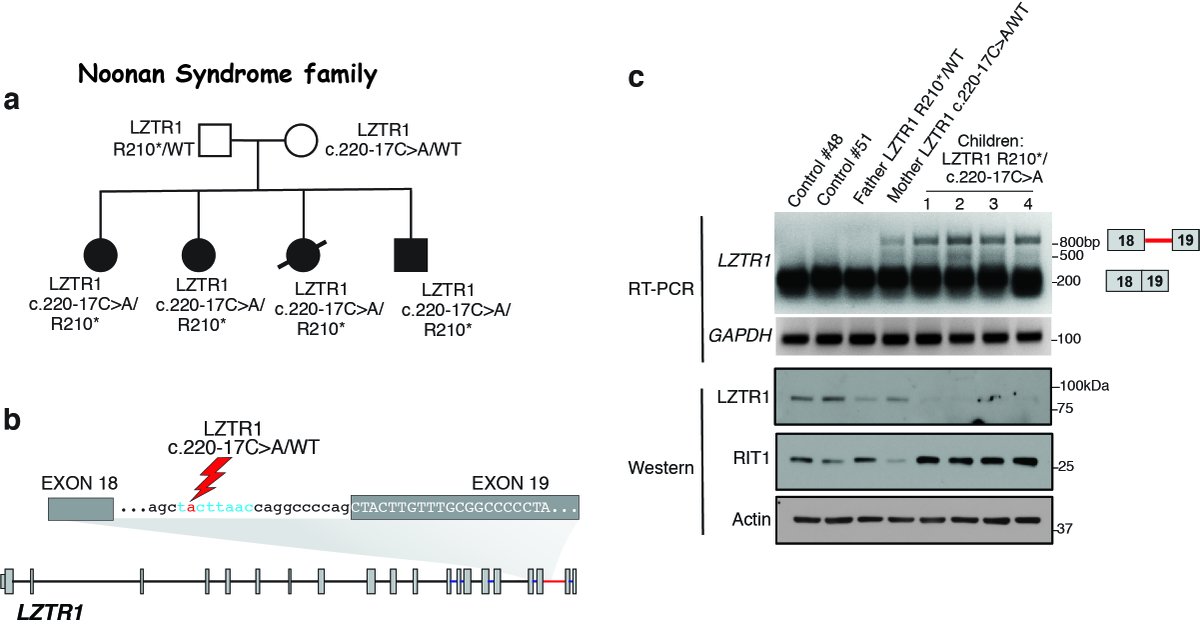
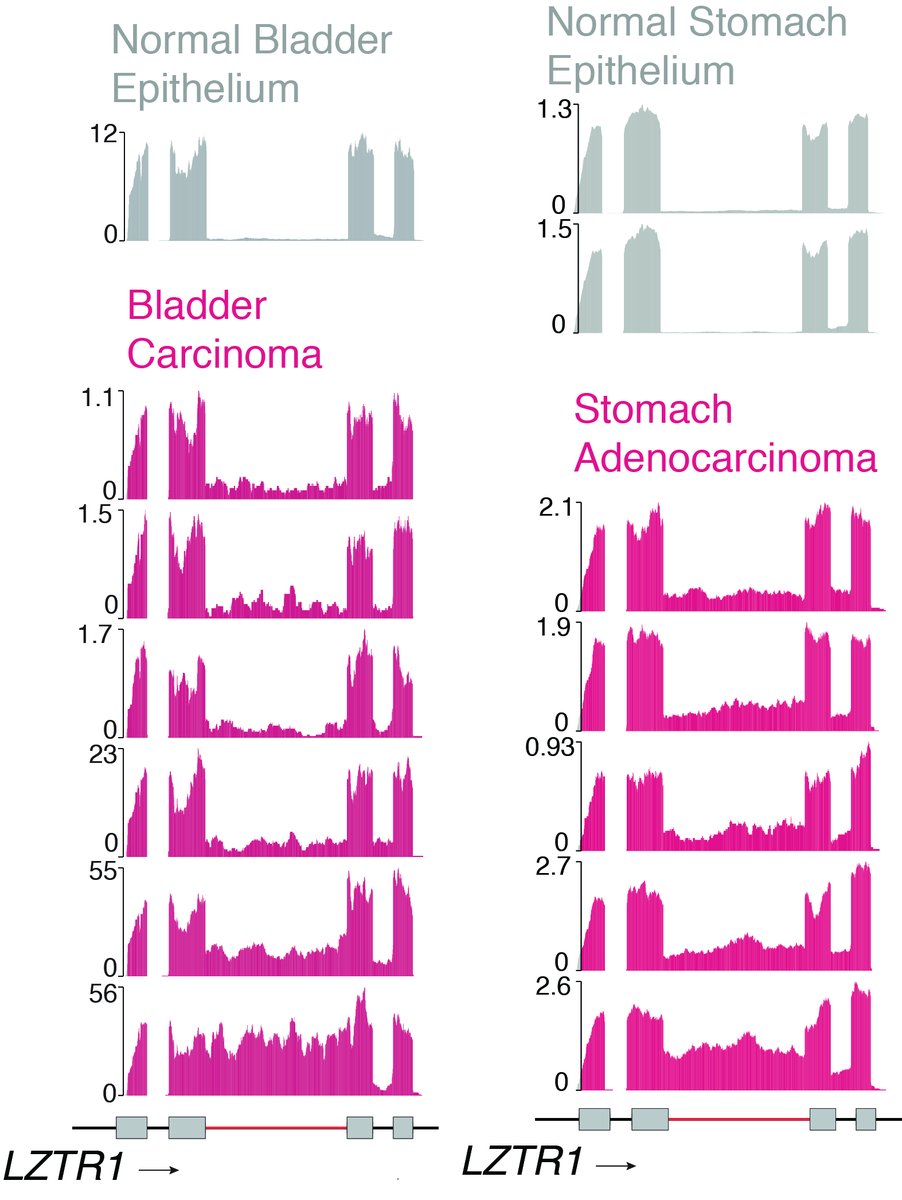
9.Moreover, in analyzing LZTR1 minor intron regulation across the pan-can #TCGA dataset, @chewomics (@csi_singapore) identified widespread impaired LZTR1 minor intron excision across cancers (the basis for which we are actively working to understand now): 
• • •
Missing some Tweet in this thread? You can try to
force a refresh









