In new study, we compared specificity of #SARSCoV2 antibody response elicited by Moderna mRNA-1273 vaccine vs infection. Some interesting differences: vaccine neut activity more RBD targeted, but has broader binding within RBD: biorxiv.org/content/10.110… (1/n)
First @AllieGreaney & Andrea Loes quantified how important RBD-binding antibodies were for neutralization by mRNA-vaccine- and infection-elicited sera. Vaccine sera neutralization was highly RBD directed: >90% of neut by nearly all vaccine sera due to RBD-binding antibodies (2/n) 

This result is interesting, as mRNA-1273 encodes entire spike ectodomain with stabilizing 2P mutations. But either those mutations or differences in antigen presentation by mRNA vaccine vs viral infection cause vaccine neut antibodies to focus more heavily on RBD. (3/n)
Note that we performed neuts w lentiviral pseudotype in ACE2-overexpressing cells, which can (over?)-emphasize importance of RBD antibodies for neutralization (nature.com/articles/s4159…). But same assay used for vaccine & convalescent sera, so difference between them is real. (4/n)
Next @AllieGreaney used deep mutational scanning developed w @tylernstarr to map how mutations in RBD affected sera antibody binding. As shown below, vaccine escape maps flatter, especially at early timepoint. Means vaccine sera binding less affected by any single mutation (5/n) 

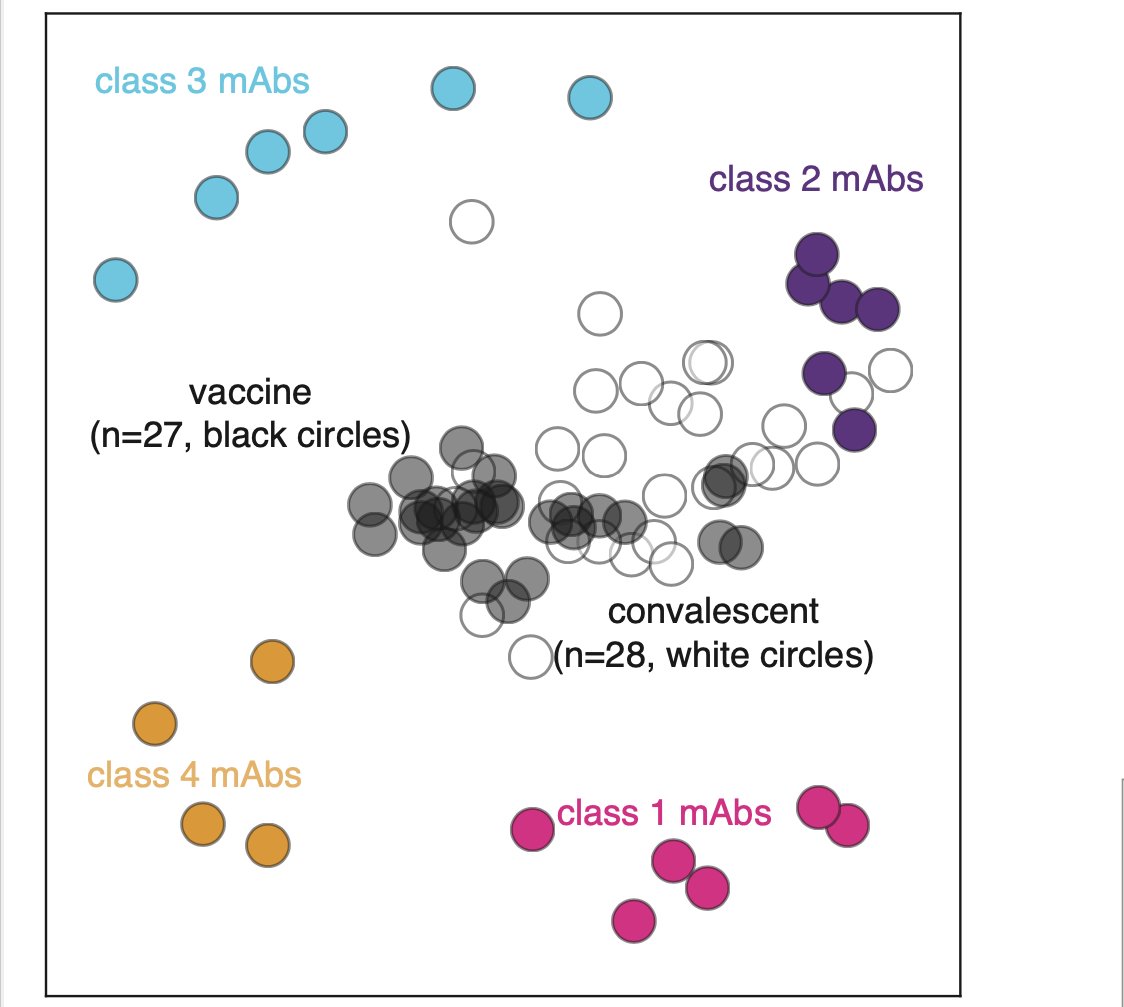
We can look at data other ways. Below is 2D projection of binding-escape from sera & monoclonal Abs in @cobarnes27 @bjorkmanlab class scheme. As reported before (biorxiv.org/content/10.110…), convalescent sera dominated by class 2 Abs, but vaccine sera more centered among Abs (6/n) 
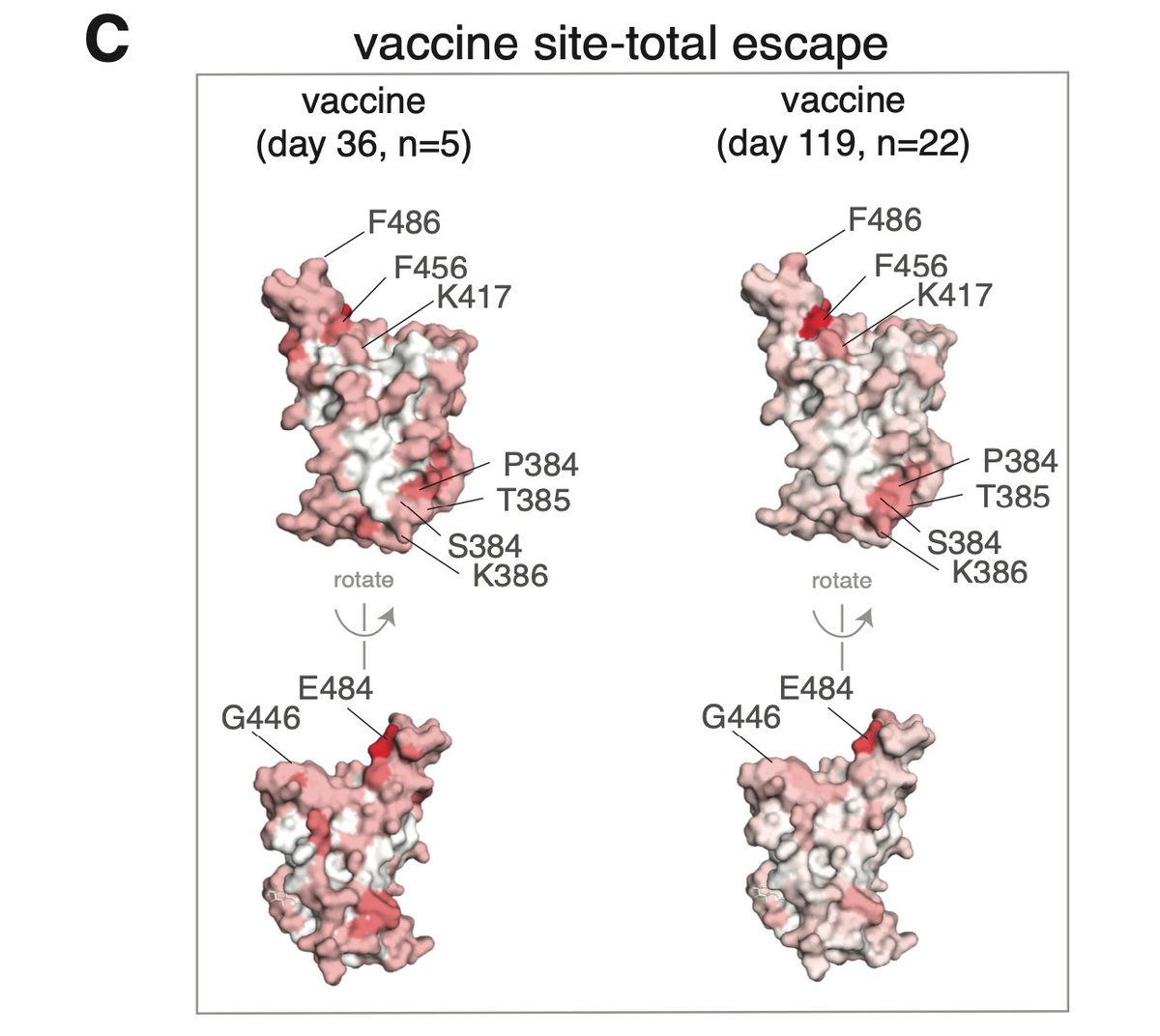
We can also look at structure. Binding escape from infection-elicited sera heavily dominated by mutations at a few sites such as E484 and F456, and also to some degree in class 3 epitope containing G446. (7/n) 

Here is what it looks like for vaccine sera. Sites E484 and F456 are also most prominent binding escape for vaccine sera, but they stand out less than for convalescent sera, again showing vaccine sera antibody binding is more "balanced" across RBD. (8/n) 
Finally, we tested key RBD muts in neut assays. As suggested by binding escape maps, single muts caused less dramatic losses in neutralization for vaccine sera.... (9/n)
... For instance, E484K reduced neut by all sera. But for some convalescent sera, E484K eliminated all RBD-directed neutralization. For vaccine sera, it took combined mutations in epitopes of class 1, 2, and 3 Abs to get comparably large decreases. (10/n) 

Overall, results indicate there are some differences in specificity of infection- & mRNA-vaccine-elicited antibodies. In particular, vaccine neutralization more RBD-focused, but this probably isn't a bad thing as binding within RBD is broader for vaccine antibodies (11/n)
Why these differences exist is question for biochemists (if due to spike 2P muts) or immunologists (if due to vaccine vs infection response), but will be interesting to understand. Also has implications for viral evolution if mutations affect different sera differently. (12/n)
Finally, study led by @AllieGreaney w important work by Andrea Loes, @LaurenEGentles, @khdcrawford, @tylernstarr, & Keara Malone. @HelenChuMD and her HAARVI cohort continue to be invaluable collaborators, & thanks to @NIAIDNews for sharing sera from phase 1 vaccine trial. (13/n)
@AllieGreaney's binding-escape mapping of Moderna vaccine sera is added to our interactive escape map viewer, as well as linked CSV if you want raw data from all studies: jbloomlab.github.io/SARS2_RBD_Ab_e… Here is screenshot of overview plot of relationships among sera & antibodies. 

• • •
Missing some Tweet in this thread? You can try to
force a refresh