Chise @sailorrooscout is a very popular account tweeting positive news about vaccines.
Unfortunately, she usually omits all caveats balanced reviews of the studies contain.
I therefore recommend everyone to read her tweets with a big grain of salt. Here's a case study thread.
Unfortunately, she usually omits all caveats balanced reviews of the studies contain.
I therefore recommend everyone to read her tweets with a big grain of salt. Here's a case study thread.
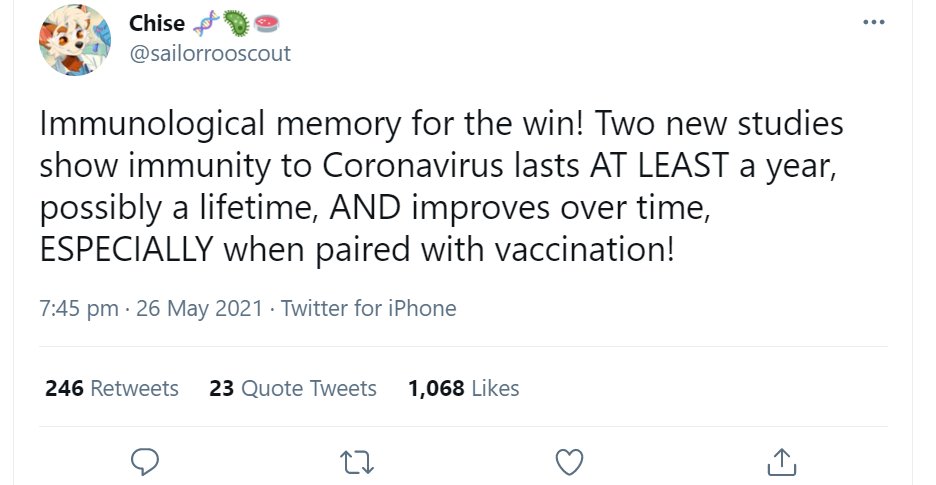
Today, Chise tweeted the following positive news.
Before I looking at her thread in detail, let's first have a look at what the underlying study says. I will use the excellent @nytimes article referenced by Chise as my source: nytimes.com/2021/05/26/hea…
Before I looking at her thread in detail, let's first have a look at what the underlying study says. I will use the excellent @nytimes article referenced by Chise as my source: nytimes.com/2021/05/26/hea…
The headline contains a really good, balanced summary:
"Immunity to the Coronavirus May Persist for Years, Scientists Find
Important immune cells survive in the bone marrow of people who were infected with the virus or were inoculated against it, new research suggests."
"Immunity to the Coronavirus May Persist for Years, Scientists Find
Important immune cells survive in the bone marrow of people who were infected with the virus or were inoculated against it, new research suggests."

Chise's first tweet is much less clear, swapping out accuracy for optimism.
Note that the NYT writes: "may persist for years".
Chise turns that into a categorical claim: "immunity lasts AT LEAST a year"
Note that the NYT writes: "may persist for years".
Chise turns that into a categorical claim: "immunity lasts AT LEAST a year"

The rest of her short thread paraphrases two paragraphs from the article.
But what are the caveats? Chise mentions none. Even though the article does. So let's have a look what Chise omitted other than the word "may".

But what are the caveats? Chise mentions none. Even though the article does. So let's have a look what Chise omitted other than the word "may".


The study suggests that for most people with previous infections one dose of a vaccine will be enough.
However, the article mentions that there were still some people who did not have a robust response after infection and should still get two doses (in theory).
However, the article mentions that there were still some people who did not have a robust response after infection and should still get two doses (in theory).

The good news is that B cells keep maturing and last for a long time. It seems that repetitive infections with other coronaviruses occur mostly not because of waning immunity but because these viruses mutate and there escape existing immunity. 

Note: this is exactly what seems to happen with Sars-CoV-2. Vaccinations still work well against B.1.617.2, for instance, but the effectiveness is somewhat reduced.
So while immunity to a particular virus may last for years, the virus may mutate and therefore evade.
So while immunity to a particular virus may last for years, the virus may mutate and therefore evade.
It appears as though immunity after infection with the real virus is better compared with vaccination alone. Not surprising, after all, the vaccines often just contain a single part of the virus and often don't remain in the body for as long. 

Chive left out this very important caveat:
5 out of 19 patients did NOT have detectable B cells in their bone marrow, despite having been infected previously. Despite what Chise's optimism may make one think, immunity is never going to be perfect for everyone.
5 out of 19 patients did NOT have detectable B cells in their bone marrow, despite having been infected previously. Despite what Chise's optimism may make one think, immunity is never going to be perfect for everyone.

B cells kept maturing, working better against variants over time. But it's important to note that neutralising activity against variants like B.1.351 was still less than for the wildtype. 

The last paragraph is very important: it warns against generalising the study's results to people who were only vaccinated and had not previously been infected. 

Those are my takeaways from the article. It's good news, but I don't feel Chise communicates the results well. What do you think now that you've seen both presentations? What's the feeling you get when you read Chise's threads? Is it just me who feels they are overly optimistic?
• • •
Missing some Tweet in this thread? You can try to
force a refresh








