
Studying SARS-CoV-2 evolution @nextstrain @biozentrum
Studied Physics @Cambridge_Uni, then learned about computers
@corneliusroemer.bsky.social
16 subscribers
How to get URL link on X (Twitter) App


 In my very rough reading, there's not enough data yet to pin down growth advantage. It could be small or non-existent, or it could be sizeable, e.g. doubling every week.
In my very rough reading, there's not enough data yet to pin down growth advantage. It could be small or non-existent, or it could be sizeable, e.g. doubling every week.
https://twitter.com/nextstrain/status/1689965962852708352EG.5 is one of the fastest growing XBB sublineages, particularly common in China where it appears to be dominant.




 BQ.1.1 and XBB have quite a lot of spike differences and seem to have similar growth advantages - this makes them candidates for co-circulation.
BQ.1.1 and XBB have quite a lot of spike differences and seem to have similar growth advantages - this makes them candidates for co-circulation.
https://twitter.com/ViolaPriesemann/status/1578300220764753922
 In der Veröffentlichung & zugehöriger Pressemeldung steht, dass es momentan nicht möglich sei zu prognostizieren welches Szenario eintritt.
In der Veröffentlichung & zugehöriger Pressemeldung steht, dass es momentan nicht möglich sei zu prognostizieren welches Szenario eintritt.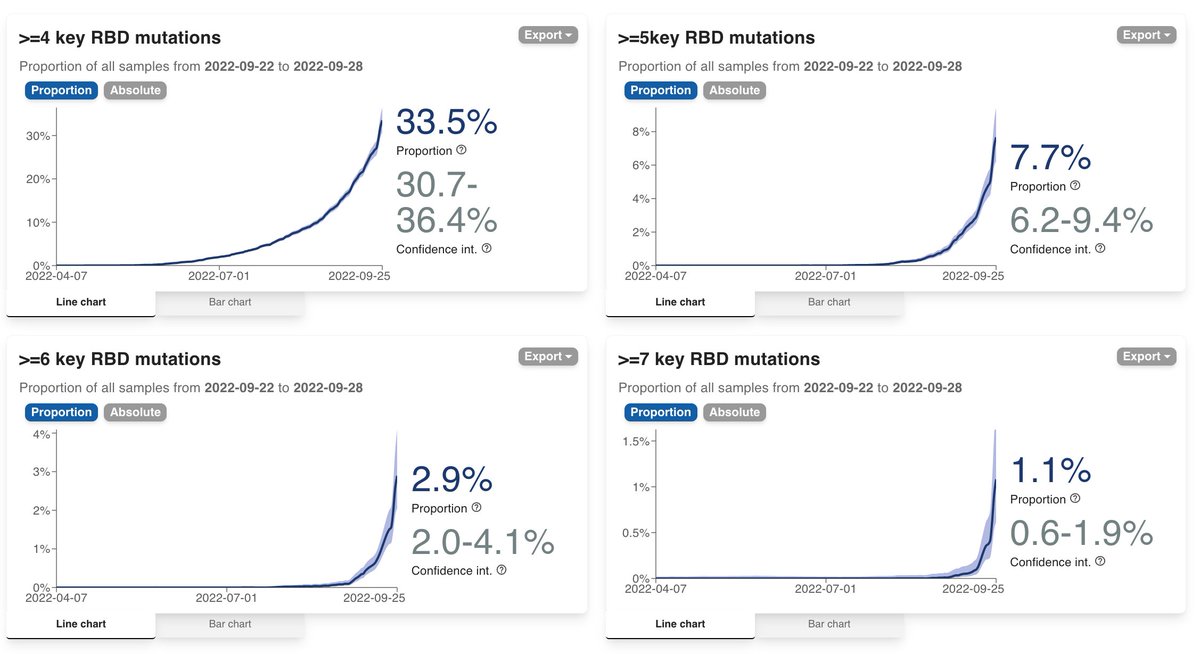
 Looking at the data like this, we reduce the complexity behind numerous emerging variants into 4 buckets of increasing escape. This is not perfect as what's in a bucket is not totally the same, but it solves the problem of having to remember BQ.1.1, BA.2.75.2, XBB etc. 2/
Looking at the data like this, we reduce the complexity behind numerous emerging variants into 4 buckets of increasing escape. This is not perfect as what's in a bucket is not totally the same, but it solves the problem of having to remember BQ.1.1, BA.2.75.2, XBB etc. 2/ 

 In the example above, for the US, BA.2.75* lineages make up 40%, BQ.1 makes up 30%, and other lineages make up the other 30%.
In the example above, for the US, BA.2.75* lineages make up 40%, BQ.1 makes up 30%, and other lineages make up the other 30%.
https://twitter.com/CorneliusRoemer/status/1572520978433060870
 A week ago, @TWenseleers estimated a growth advantage of ~14% per day for BQ.1 over BA.5
A week ago, @TWenseleers estimated a growth advantage of ~14% per day for BQ.1 over BA.5https://twitter.com/TWenseleers/status/1574070423175966720?s=20&t=DwpPGSXkUpGF7lfeLXhMYw



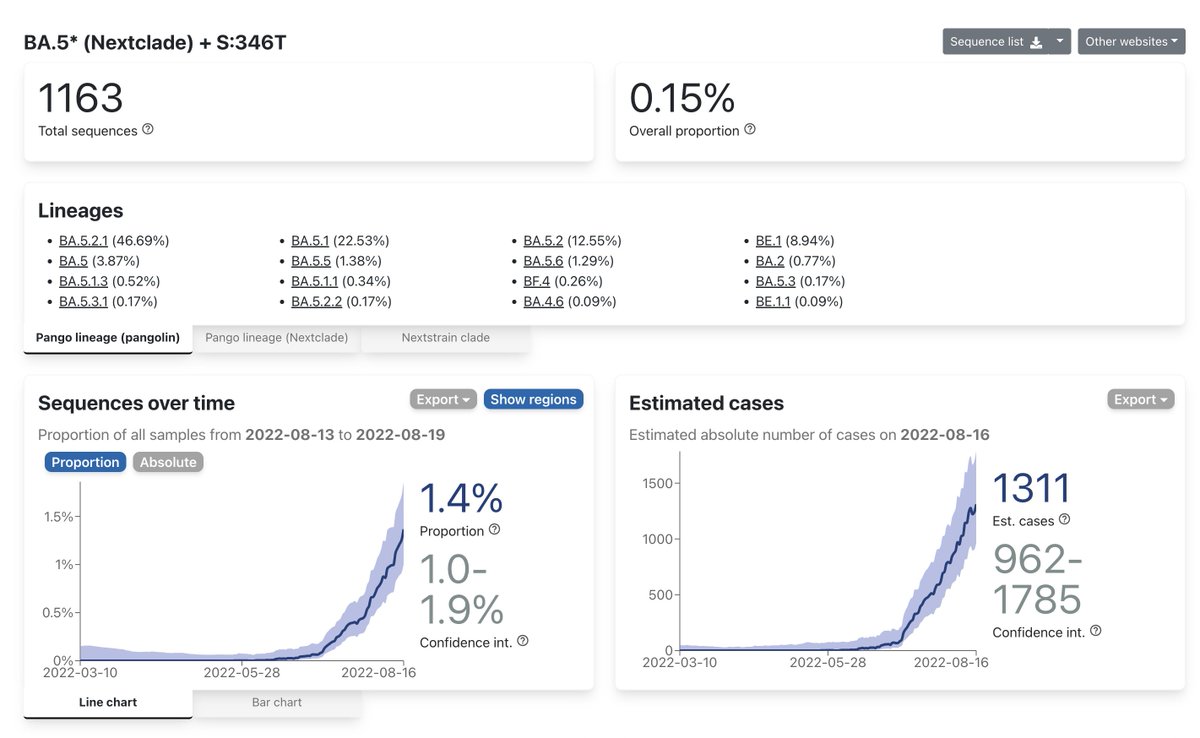
 BA.4.6 has slowed in growth because BA.5 + S:346T is fitter than BA.4 + S:346T (we have much more BA.5 now than BA.4)
BA.4.6 has slowed in growth because BA.5 + S:346T is fitter than BA.4 + S:346T (we have much more BA.5 now than BA.4)
https://twitter.com/ViolaPriesemann/status/1567135669570387969Wie häufig haben Sie sich PCR-bestätigt infiziert?

https://twitter.com/TWenseleers/status/1561374092607033350Various BA.2.75 lineages have already acquired a variety of RBD mutations e.g. at S:486, S:490, S:452

 I'm showing Germany and the US here because they publish the most consistent and smooth data.
I'm showing Germany and the US here because they publish the most consistent and smooth data.
https://twitter.com/nextstrain/status/1550615442695151616When the Pango committee gave the names BA.4/5 a name BA.2.X would have been also justifiable.


 Hier ein Faktencheck der Aussagen.
Hier ein Faktencheck der Aussagen.
https://twitter.com/caceypeters/status/1547953624461938688So many amazing videos there, thanks @caceypeters
https://twitter.com/caceypeters/status/1297013036646125568?t=6gVm_DB9msQ9yKOBYHJYwQ&s=19