
Proteins that interact together are often relevant for the same traits and network analyses of GWAS genes can help identify trait relevant cell biology. Here @ibarrioh applied this to study 1002 traits defining a pleiotropy map of human cell biology
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
GWAS genes were selected with the locus to gene score which incorporates diverse features ( genetics-docs.opentargets.org/our-approach/p…) and a comprehensive network was assembled by @intact_project Network expansion can recover disease genes and drug targets not found by GWAS 

The similarity of the network expansion scores finds human traits with common biological basis and the biological processes that underlie this. We find 73 gene modules linked to >1 trait including a group of highly pleyotropic gene modules (e.g. general RNA/protein regulation) 

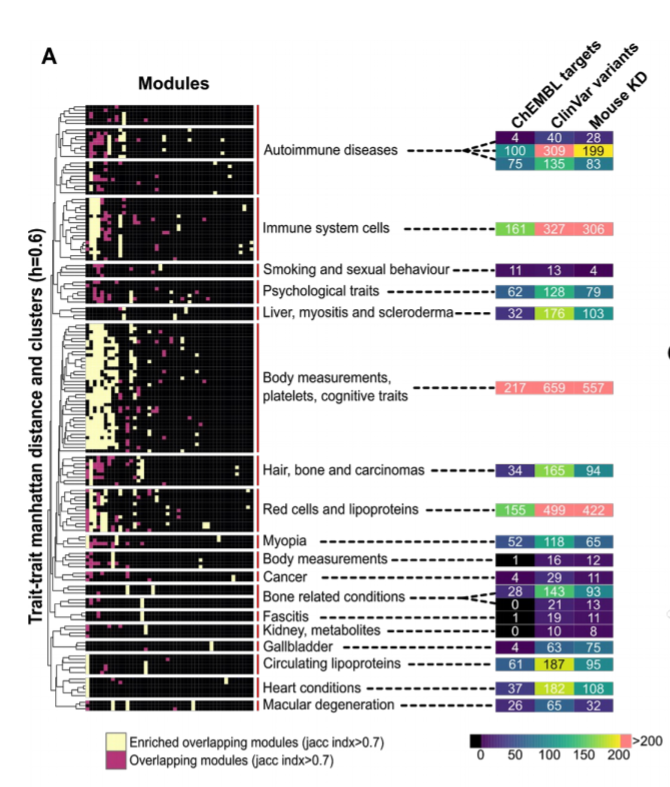
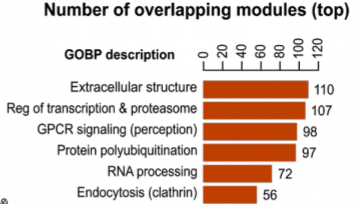
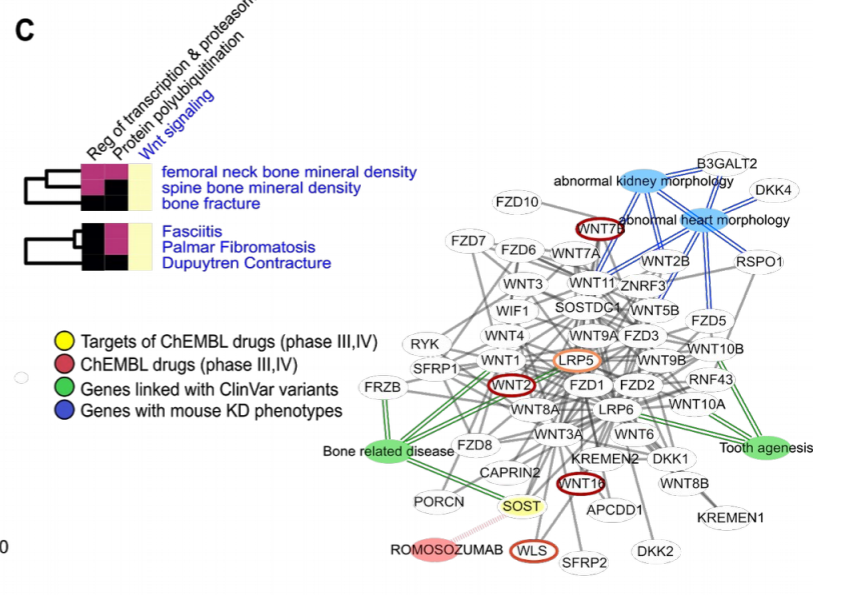
We find examples of gene modules linked to groups of related traits that are also enriched in protein carrying disease variants. These disease related variants from patients are often not found on GWAS linked genes even if within the same process. 
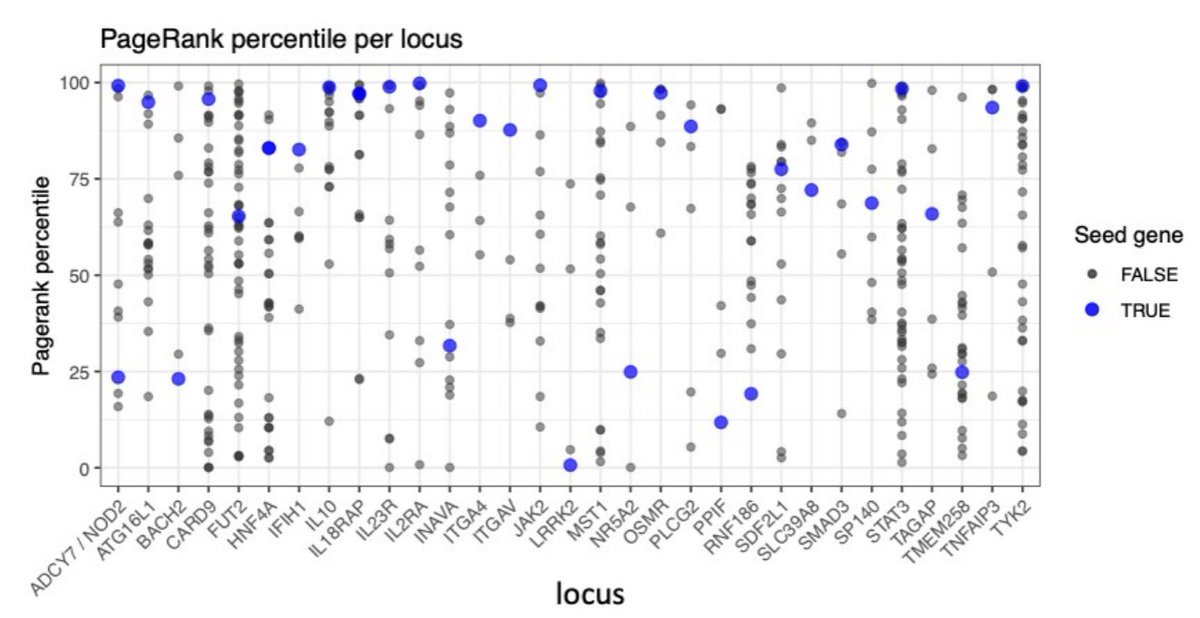
Finally, with @jschwart37 and @anderson_carl we illustrate the value of the network expansion scores to help prioritise candidate genes in trait associated loci for IBD diseases and we provide a list of genes with strong functional and genetic support. 
This was a collaboration between many groups involved in Open Targets opentargets.org. I find that there is some isolation between those that speak GWAS, cell biology and structural biology and real benefit in having more such collaborations.
For GWAS people, there is a somewhat dogmatic avoidance of prior knowledge before the associations are done while I feel that sometimes it is worth trading off causality and some aspects of unbiased analysis for clarity and mechanisms.
• • •
Missing some Tweet in this thread? You can try to
force a refresh






