Anyone want to guess what’s wrong with this paper?
It’s making the rounds as some reason to wear masks. academic.oup.com/cid/advance-ar…
It’s making the rounds as some reason to wear masks. academic.oup.com/cid/advance-ar…
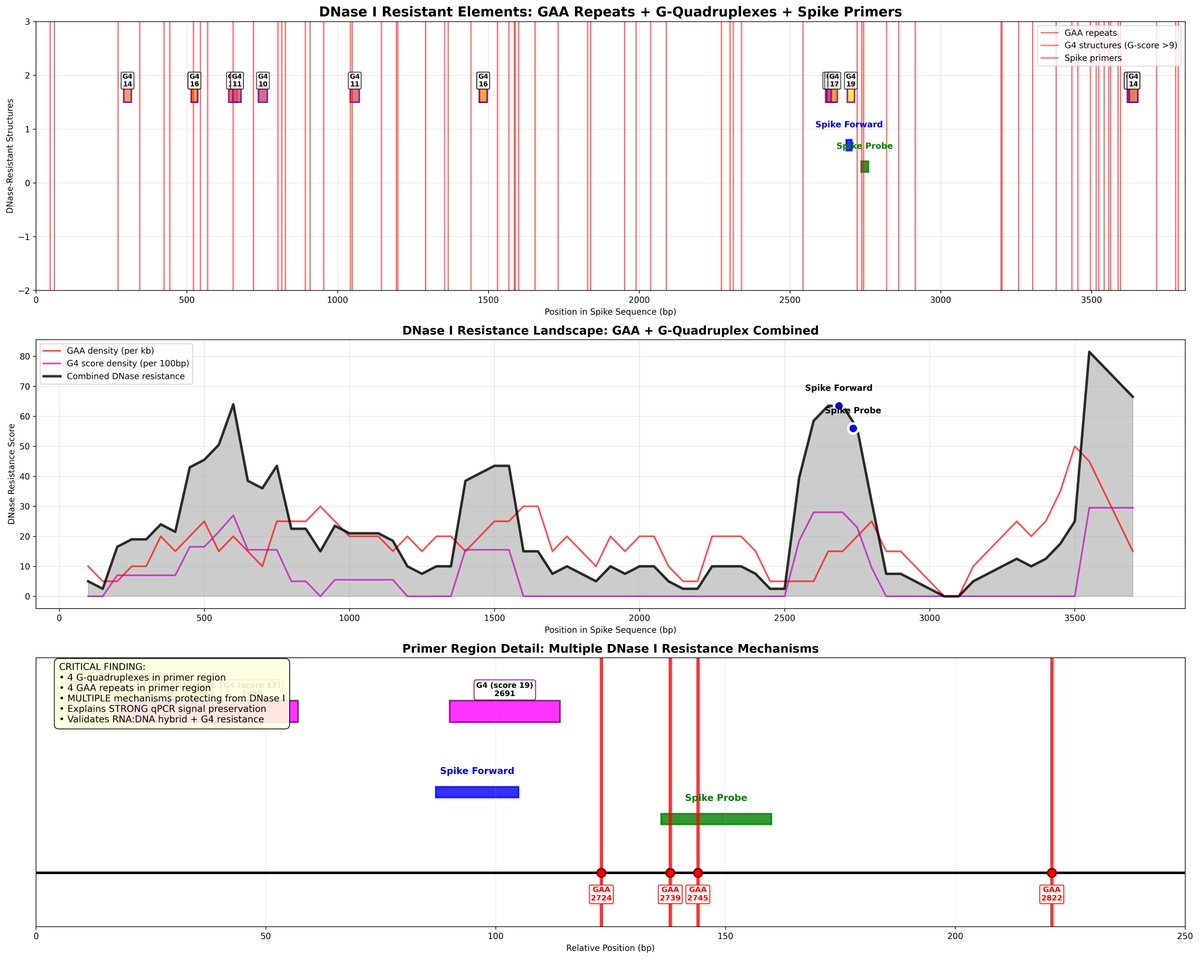
They target the N gene which is the highest copy subgenomic RNA in the genome.
SgRNA isn’t infectious.
N numbers are tiny.
SgRNA isn’t infectious.
N numbers are tiny.

Orders of magnitude.
This is pure Merchants of Mask Malaise.
Fear porn, spun with a veil of credibility and timed for school.
This is pure Merchants of Mask Malaise.
Fear porn, spun with a veil of credibility and timed for school.
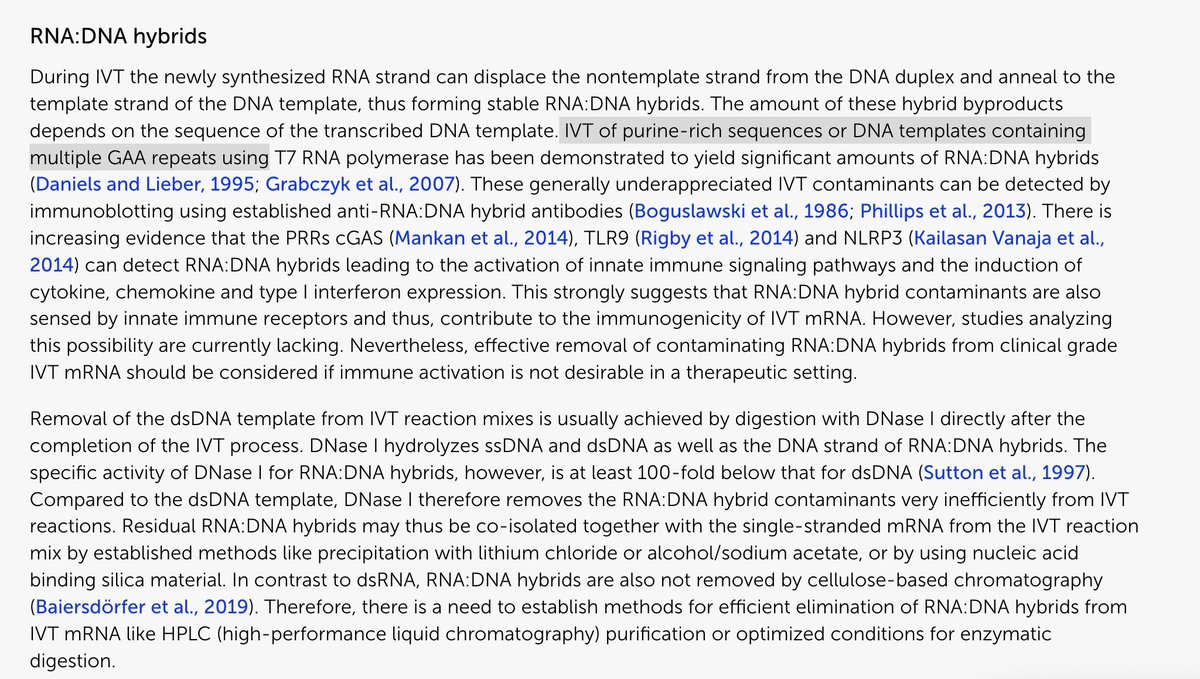
There are methods to perform viability PCR on SARs-CoV-2.
I wonder why the authors didn’t pursue those?
epa.gov/healthresearch…
I wonder why the authors didn’t pursue those?
epa.gov/healthresearch…
Just to clarify.
This is the Nanopore coverage Map.
Lowest point in the trough in the center looks like 2,000X coverage.
Highest point on the right is ~500,000X coverage.
Obviously full length infective virus can’t be more than the 2,000X and the stuff on the right is non-infect
This is the Nanopore coverage Map.
Lowest point in the trough in the center looks like 2,000X coverage.
Highest point on the right is ~500,000X coverage.
Obviously full length infective virus can’t be more than the 2,000X and the stuff on the right is non-infect

Non-infectious fragments of RNA known as sub genomic RNA or sgRNA.
Why would you place your PCR assay on the yellow region? You know you are predominately amplifying non-infectious RNA.
Deliberate witch hunting.
Why would you place your PCR assay on the yellow region? You know you are predominately amplifying non-infectious RNA.
Deliberate witch hunting.
Should could target ORF1a (in the trough of the coverage map) that makes less subgenomic RNA but her sensitivity would drop ~200 fold.
But she’s only working with 6-5,800 copies.
Most of the data would be blank.
They’d have to speak for hours...
Into a vacuum tube.
Redonkulus
But she’s only working with 6-5,800 copies.
Most of the data would be blank.
They’d have to speak for hours...
Into a vacuum tube.
Redonkulus

Interesting detail.
They did not Try to culture the coarse fraction (Over 5um).
That would have been very valuable information.
The diagnostic PCR does have low Ct but that’s PCR with diff assay/ diff lab/ diff time.
130L/min vacuum for 15-30 minutes.
They did not Try to culture the coarse fraction (Over 5um).
That would have been very valuable information.
The diagnostic PCR does have low Ct but that’s PCR with diff assay/ diff lab/ diff time.
130L/min vacuum for 15-30 minutes.

Really shouldn’t say there is anything wrong with the paper....just seeing people use this as evidence for masking kids and it does not follow.
• • •
Missing some Tweet in this thread? You can try to
force a refresh