Excited to share our new preprint on "Assessment of 3D MINFLUX data for quantitative structural biology in cells"
with @AlistairCurd
biorxiv.org/content/10.110…
We thank @LSchermelleh @HohlbeinLab @michellepeckham for the feedback and looking forwards to yours!
#MINFLUX
with @AlistairCurd
biorxiv.org/content/10.110…
We thank @LSchermelleh @HohlbeinLab @michellepeckham for the feedback and looking forwards to yours!
#MINFLUX

Thanks, @Stefan_W_Hell for sharing the data which helped to revisit the claims made in the following article "MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells" @naturemethods
nature.com/articles/s4159…
Hope our findings are useful for MINFLUX research!
nature.com/articles/s4159…
Hope our findings are useful for MINFLUX research!
First, using PERPL, our localization precision estimates of 0.98±0.02 nm, 3.20±0.05 nm and 3.31±0.08 nm broadly agreed with the published analysis but implying FWHM of 2.4-7.1 nm.
So, what should be the best theoretical resolution? 1-3 nm as claimed?
So, what should be the best theoretical resolution? 1-3 nm as claimed?

Next, we probed if MINFLUX can clearly resolve the 8-fold symmetry of Nup96 at the single pore level?
We segment 20 (~ max possible across different datasets) individual Nup96 complexes and rarely observed 8-fold symmetry.
All segmented NPCs will be available via Zenodo.
We segment 20 (~ max possible across different datasets) individual Nup96 complexes and rarely observed 8-fold symmetry.
All segmented NPCs will be available via Zenodo.

We found Nup96 diameter distributions to be 107±10 nm, 108±7 nm and 111±5 nm for 2D, 3D 1-color and 3D 2-color datasets.
Nup96 ring diameter was rarely 107 nm as referenced in Thevathasan et al.
Surprisingly, the diameter spread for MINFLUX Nup96 datasets was not unreported.
Nup96 ring diameter was rarely 107 nm as referenced in Thevathasan et al.
Surprisingly, the diameter spread for MINFLUX Nup96 datasets was not unreported.

We also found the spread in the number of localisations per pore across different datasets very interesting. 

Experimental data is always incomplete!
So next, we used PERPL, a structure-based modelling technique designed for incomplete data and ensemble analysis
We observed different symmetries for different datasets
1. 9-fold - 2D, 1-color
2. 7-fold- 3D, 1-color
3. 8-fold- 3D, 2-color
So next, we used PERPL, a structure-based modelling technique designed for incomplete data and ensemble analysis
We observed different symmetries for different datasets
1. 9-fold - 2D, 1-color
2. 7-fold- 3D, 1-color
3. 8-fold- 3D, 2-color
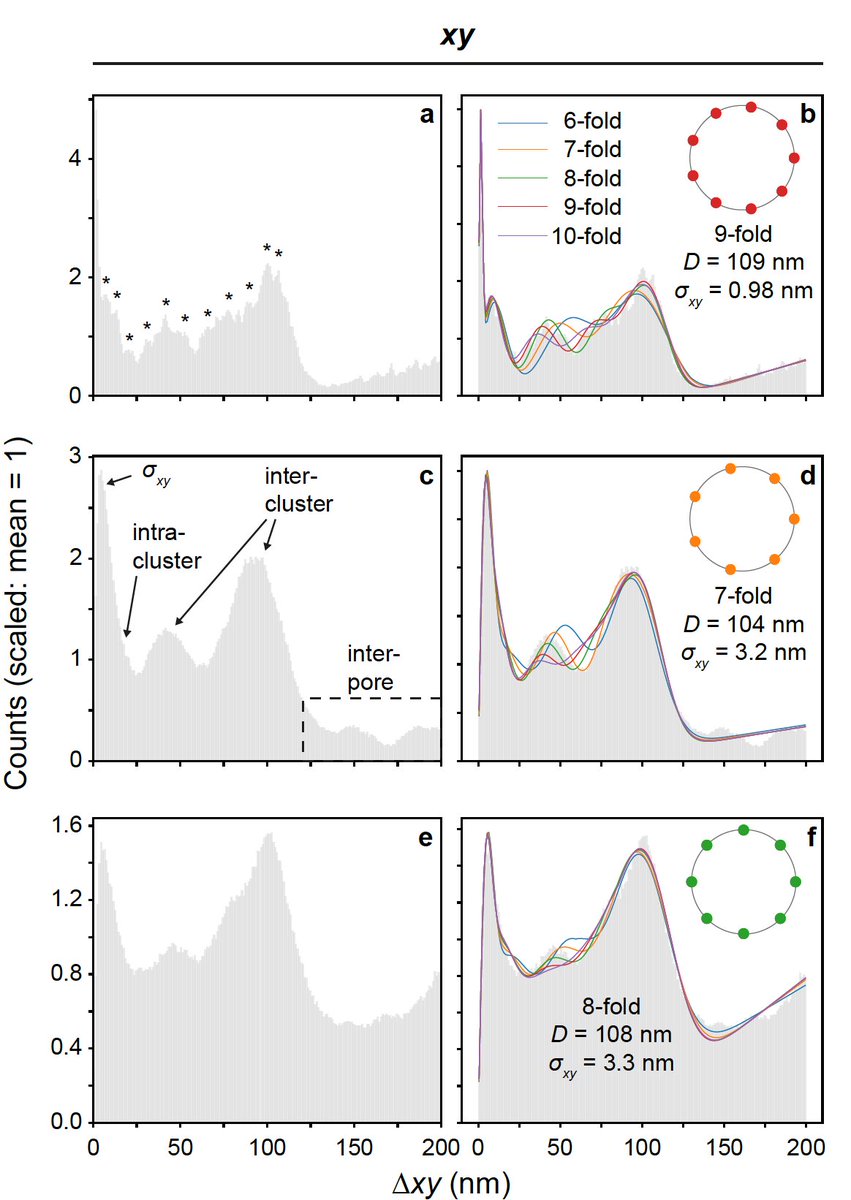
In FOV ensemble analysis, the estimates of the Nup96 ring diameters again varied from 104 to 109 nm.
Secondary peaks may indicate substructure within clusters in the 2D dataset down to a distance of 7 nm. However, this detail is lost in the 3D 1-color and 2-color datasets.
Secondary peaks may indicate substructure within clusters in the 2D dataset down to a distance of 7 nm. However, this detail is lost in the 3D 1-color and 2-color datasets.
Most surprising was the average z-distance between cyto- and nucleoplasmic layers of Nup96 localisations at 40.5 nm instead of the expected ~50 nm.
In contrast, for the 2-color dataset, the inter-layer distance was 50 nm close to the expected, the authors estimated ∼46 nm.
In contrast, for the 2-color dataset, the inter-layer distance was 50 nm close to the expected, the authors estimated ∼46 nm.

Another interesting finding was Z localisation precision of 2.28 nm and 3.08 nm for the 3D, 1-color and 2-color datasets.
What FWHM resolution do you expect?
Gwosch et al abstract "this true nanometer-scale resolution is obtained in three dimensions and in 2-color channels"
What FWHM resolution do you expect?
Gwosch et al abstract "this true nanometer-scale resolution is obtained in three dimensions and in 2-color channels"
Localisation filtering is now common for most single-molecule methods esp when done to up the precision game
For MINFLUX, it goes from 20% to 82% as the experimental complexity increases
Not a problem for DNA-PAINT methods where u have millions of locs but what if only 213 locs?
For MINFLUX, it goes from 20% to 82% as the experimental complexity increases
Not a problem for DNA-PAINT methods where u have millions of locs but what if only 213 locs?
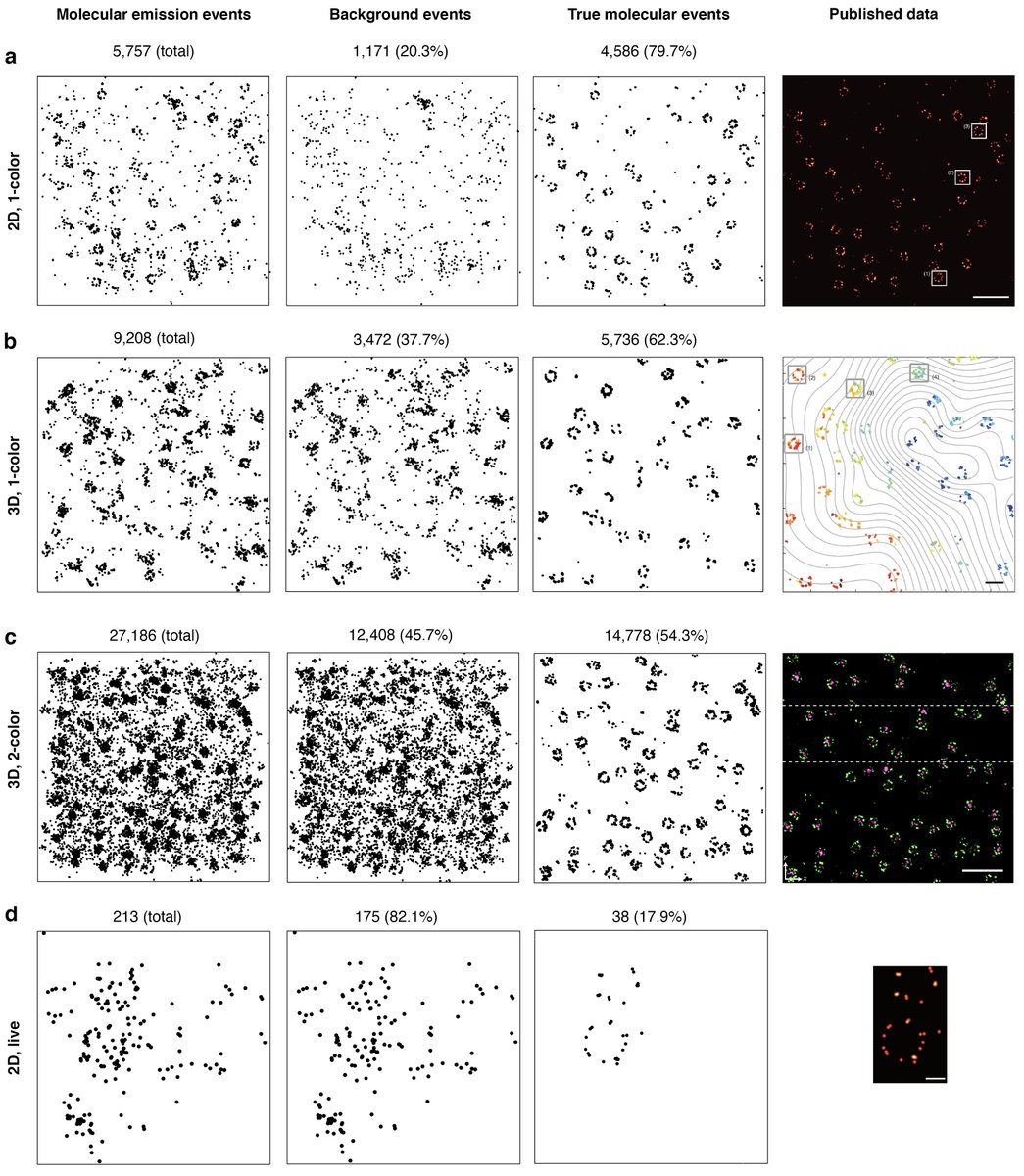
The locs filtering is most interesting when one succeeds in finding a collection of 38 localizations in form of a pore ring (live MINFLUX imaging)
How lucky😀
How lucky😀
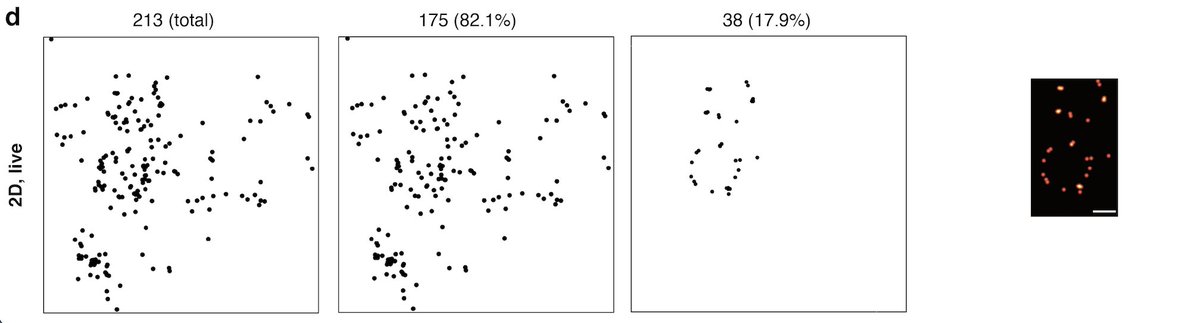
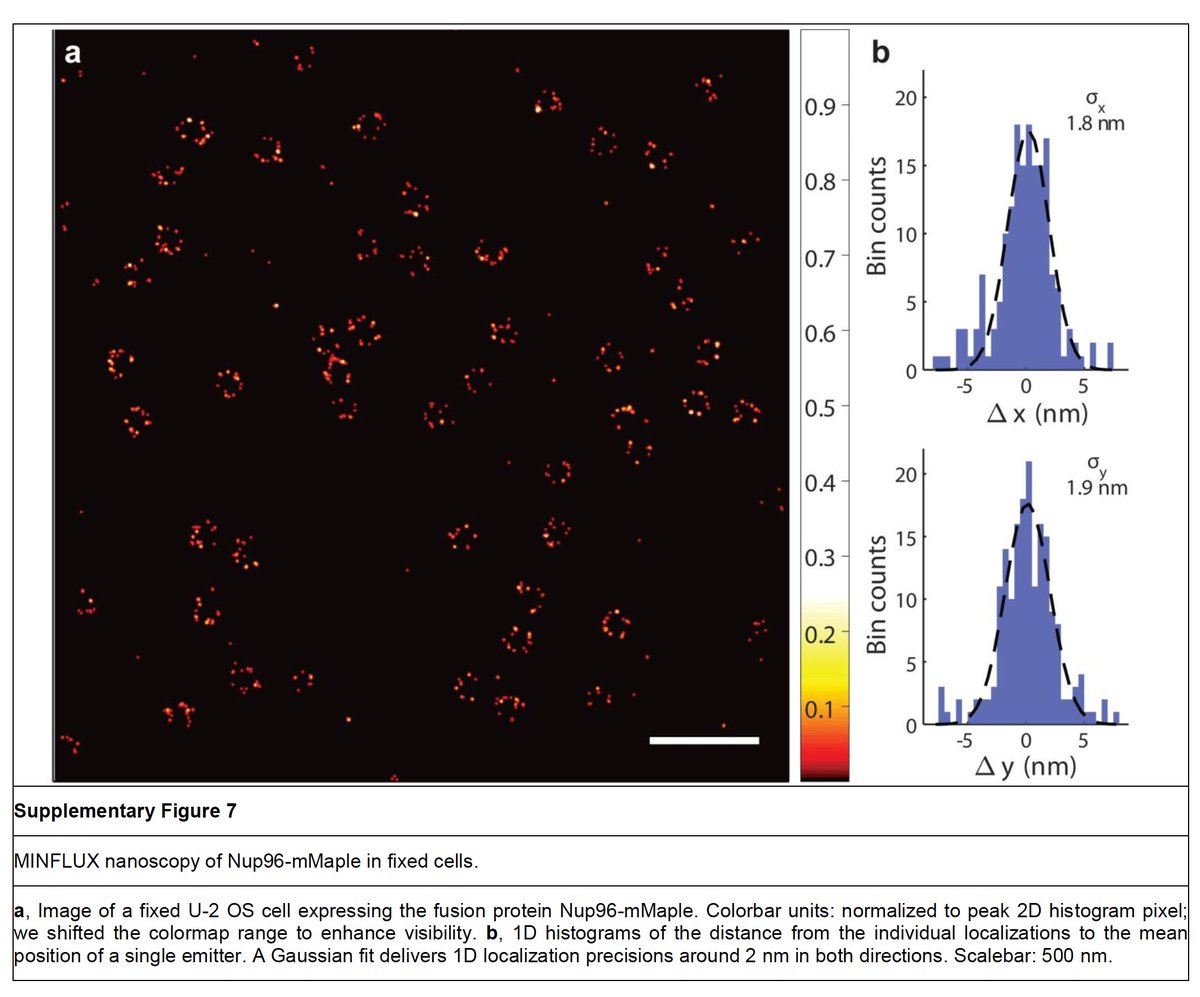
Also, remember, the measured precision ∼2 nm for Nup96-mMaple is for fixed not live samples (Sup Fig. 7 of Gwosch et al.).
To our knowledge, the localisation precision for live MINFLUX hasn't been reported.
It's all about the way you write 😀

To our knowledge, the localisation precision for live MINFLUX hasn't been reported.
It's all about the way you write 😀
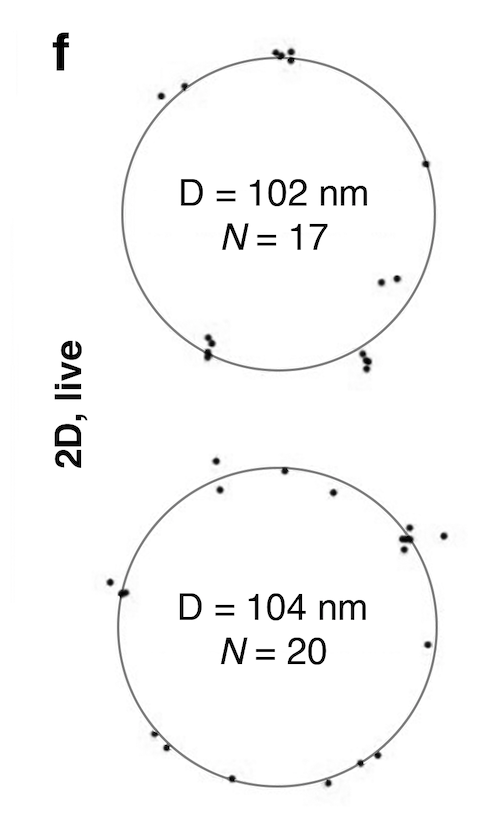
One exciting bit about live MINLUX imaging is that even with 17 and 20 localisations, one gets very close circlefit estimates for the diameter.
102 nm and 104 nm, off by only 3-5 nm from the expected 107 nm.
102 nm and 104 nm, off by only 3-5 nm from the expected 107 nm.
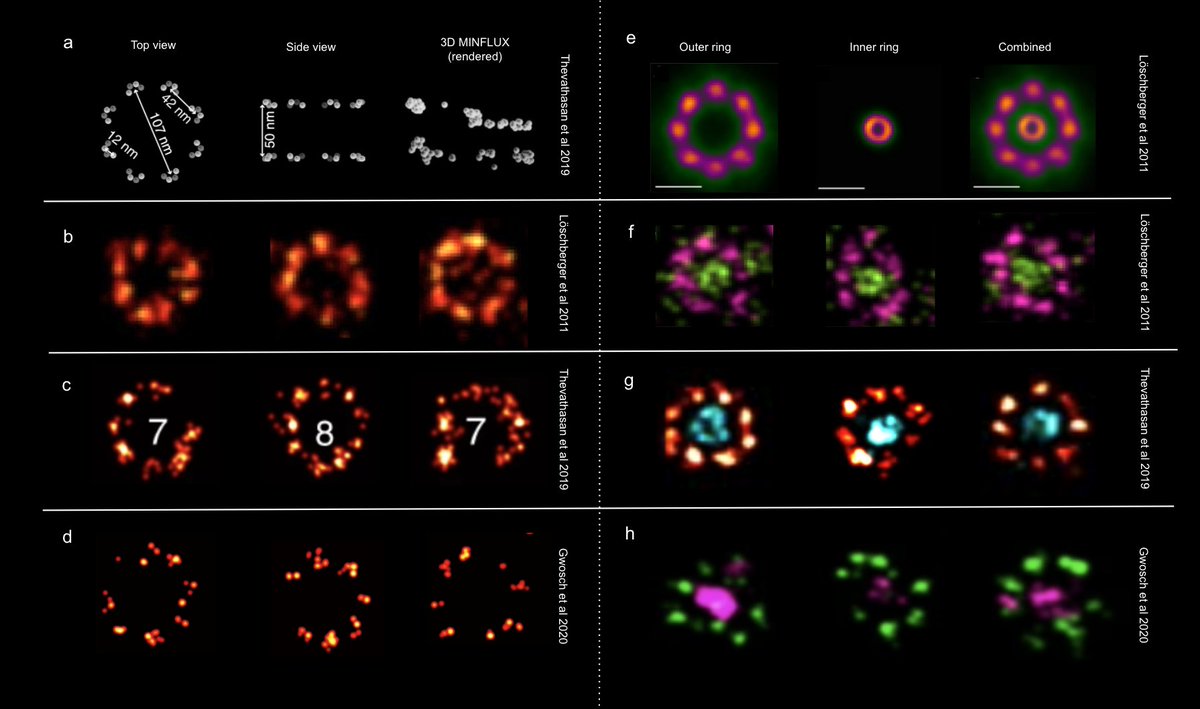
So, how does MINFLUX compare with previously published SMLM (Thevathasan et al. 2019 Nat Methods) and dSTORM (Löschberger et al. 2012 JCS) datasets?
The inner WGA ring is easily visible with SMLM/dSTORM but appears as a blob in the case of MINFLUX
The inner WGA ring is easily visible with SMLM/dSTORM but appears as a blob in the case of MINFLUX
Here are the scatter plots showing raw localisations from 10 segmented WGA complexes from the 3D, 2-color MINFLUX dataset.
Do you observe a ring?
We will soon be depositing all datasets/codes to Zenodo.
If you want to play with Python/MATLAB codes, please email us 🙏
Do you observe a ring?
We will soon be depositing all datasets/codes to Zenodo.
If you want to play with Python/MATLAB codes, please email us 🙏

was made aware of a typo in the above tweet. The sentence should read -
Surprisingly, the diameter spread for MINFLUX Nup96 datasets was not *reported*.
Surprisingly, the diameter spread for MINFLUX Nup96 datasets was not *reported*.
• • •
Missing some Tweet in this thread? You can try to
force a refresh

















