
Very excited to share our latest preprint on how to estimate SARS-CoV-2 variant abundances in wastewater. A large joint effort with @jasmijnbaaijens, Alessandro Zulli, @IsabelOtt, @cduvallet, @BillHanage, @NathanGrubaugh, @jordan_peccia, and more 1/
medrxiv.org/content/10.110…
medrxiv.org/content/10.110…
Others have worked on this problem, but there's a fundamental issue with trying to quantify variants: the mutations that define variants are so far apart that they never appear on the same read, and often not even on the same molecule(!) in heavily degraded wastewater RNA 2/
Luckily @jasmijnbaaijens had a critical insight: this is computationally identical to RNAseq quantification! You have an unknown mixture of transcripts (known variants) which you've chopped up and noisily turned into sequencing reads, and are inferring the original mixture 3/ 

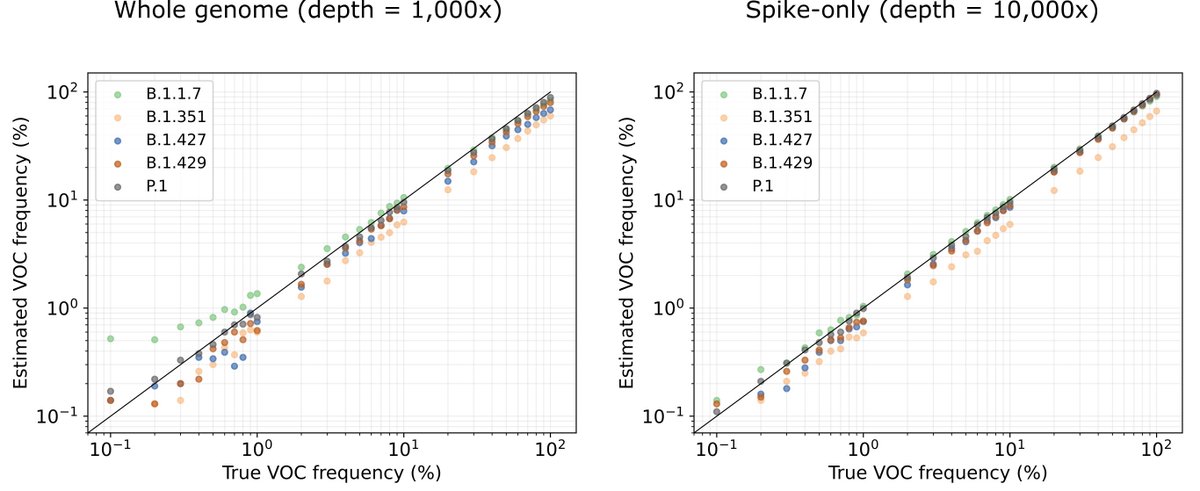
When we tested using kallisto for prediction on simulated data, it worked really, really well. There are some systematic over-and-under estimates, which speaks to the importance of choosing your reference set well, but otherwise, it works 4/ 
At the same time, Alessandro Zulli in @jordan_peccia's lab was sequencing from wastewater, and @IsabelOtt and @NathanGrubaugh's crew were looking at variants in clinical samples in the exact same area. A perfect case to try it out! 5/
It worked pretty well! Importantly, though, we found that quantification of individual wastewater samples appeared noisy, but regional trends in clinical abundance were well captured from the wastewater 6/ 

We then worked with @cduvallet and team at @BiobotAnalytics, to test our technique on samples from sixteen sites across eight states sequenced with a different workflow. Again, quite noisy, but the point is that this is a fairly general technique that works on many data types 7/ 

As you might expect, it’s important that the wastewater has low Ct for sequencing to have enough molecules to capture enough diversity to estimate variant abundance. From testing we found that while the accuracy remained decent with low coverage, precision dropped off. 8/ 
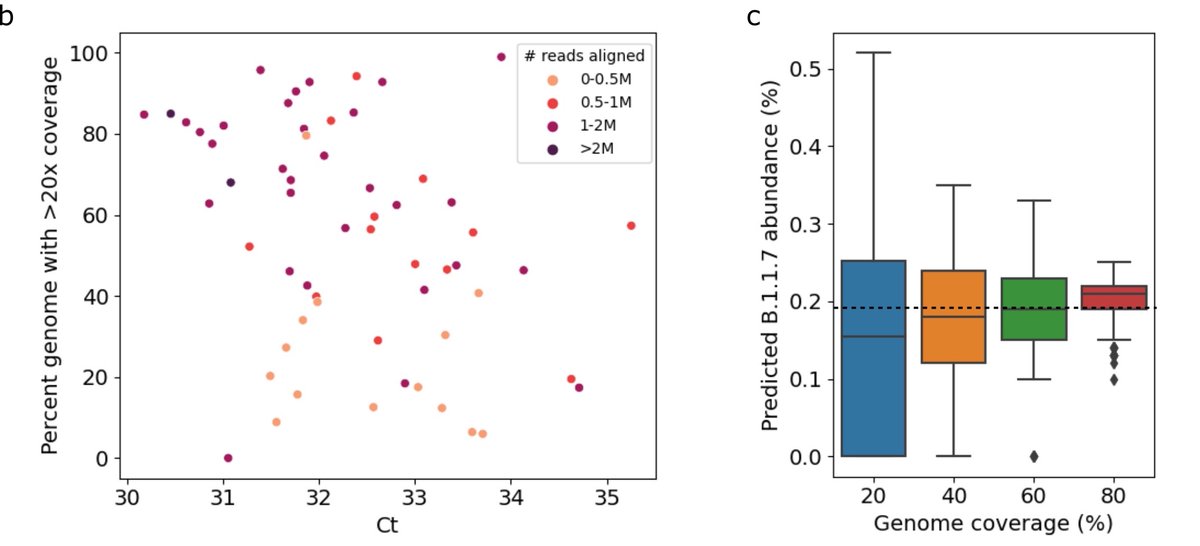
Whether the discrepancy between the wastewater and clinic is noise from the inherent noisiness of wastewater, or noise in clinical sequence data, it’s impossible to tell. But it does seem that computationally this is about as good as we’re going to do. 9/
So what do we learn from this? You can see SARS-CoV-2 variants in wastewater, but it does seem like clinical sequencing gives a more accurate and earlier picture of what’s happening. For places without robust clinical sequencing, this could help pandemic surveillance. 10/
While right now the US at least is all Delta all the time, we all know that may change again. And in any case, this is far from the last pandemic we will see, and far from the last time we will need to monitor real-time pathogen evolution on a population scale. 11/11
Coda, since it's come up: One of the key things we did to make this work was to include multiple reference sequences per variant lineage. That way instead of being thrown off by natural diversity within the lineage, that diversity actually helped make predictions even better!
• • •
Missing some Tweet in this thread? You can try to
force a refresh




