
We are very excited to share our work in @Nature on developing scRibo-seq, a method for #singlecell ribosome profiling! With @jervdberg, Amanda Andersson-Rolf, @HansClevers, and @AlexandervanOu1 nature.com/articles/s4158… 1/15
Ribosome profiling is a technique used to provide a high-resolution and instantaneous view of translation by sequencing ribosome-protected footprints
By directly integrating the footprint generation with library construction, we were able to dramatically increase the sensitivity and scalability so that we could perform ribosome profiling on single cells. 
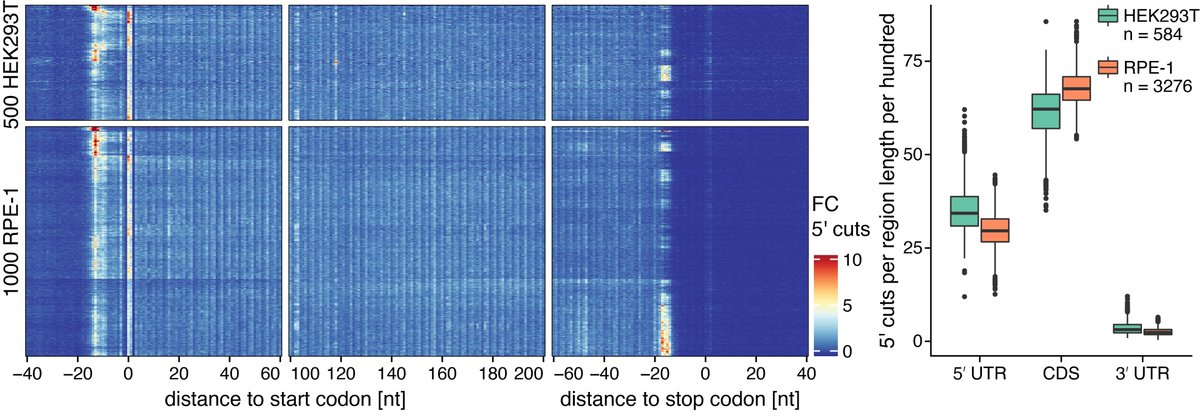
Our scRibo-seq libraries exhibit the same characteristics of bulk Ribo-seq, with reads predominantly aligning to coding sequences, an increase in local read density over the start and stop codons, and a 3-nucleotide periodicity in how the reads map to the CDSs. 
We further validated that we were indeed measuring translation by applying scRibo-seq to cells that had been starved of arginine or leucine. As expected, the starvation treatments caused an increase in footprint density over a subset of the codons encoding the removed amino acids 
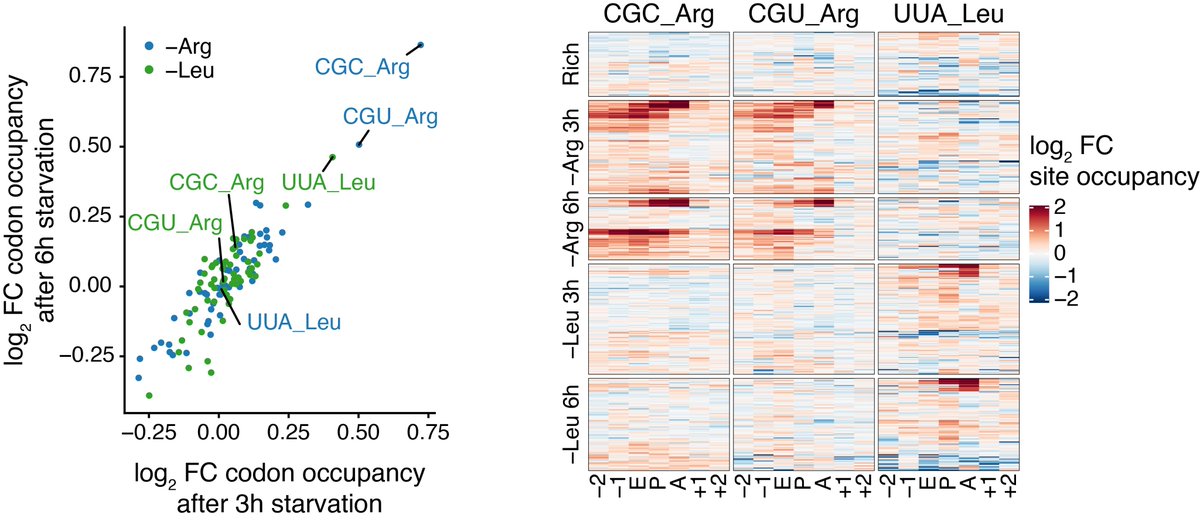
Interestingly, the response to amino acid starvation was not uniform across the population of cells. We saw that those with the largest ribosome pausing response to arginine (CGC & CGU) starvation were also translating histone genes. 
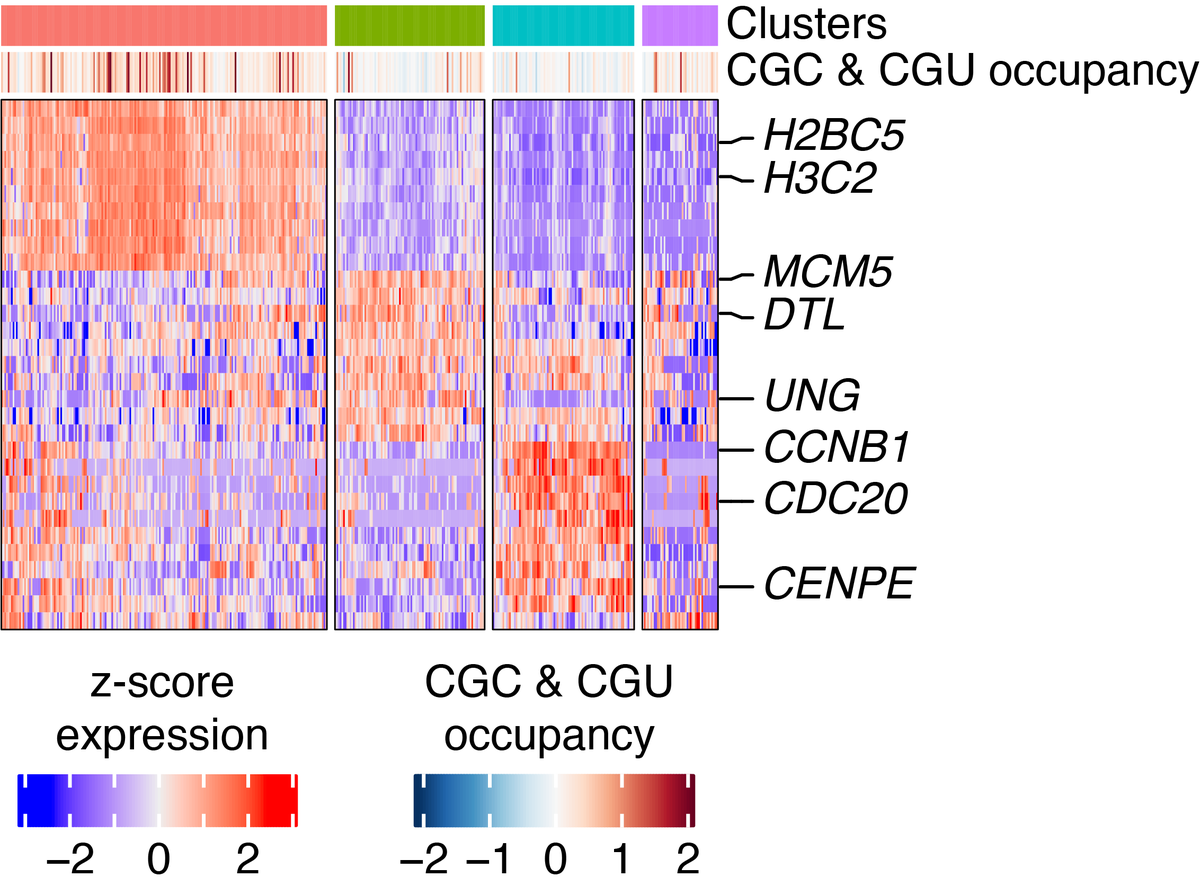
As histones are only expressed during S phase, the cell cycle thus has an influence on the effect of amino acid limitation on translational pausing.
Having seen this cell-cycle effect, we next wondered if translational properties changed through the unperturbed cell cycle. We thus applied scRibo-seq to RPE-1 FUCCI cells collected from different cell fractions to ensure the entire cell cycle was represented.
Cluster analysis and pseudotime ordering of these cells revealed not only the expected change in abundances of known cell-cycle marker genes, but also that the frequency of certain codons in the ribosome footprints also varies. 
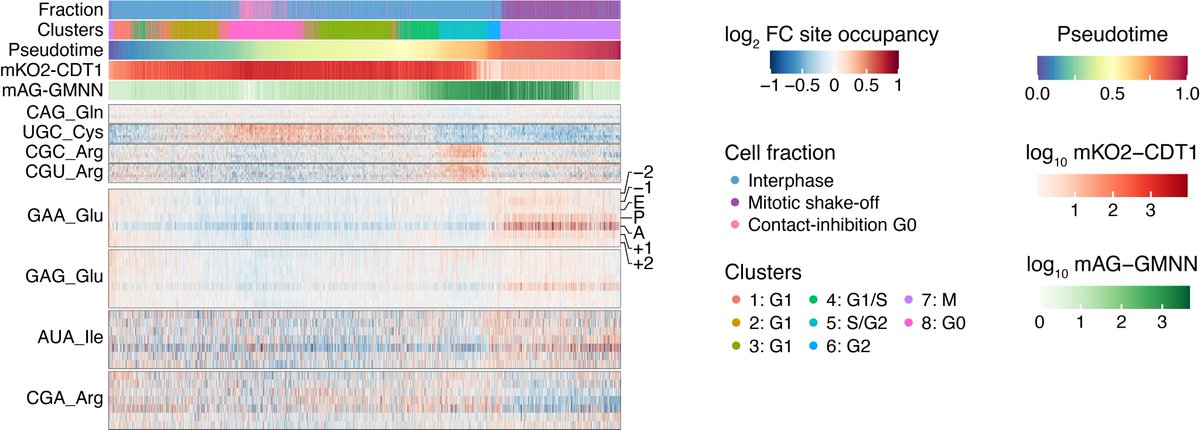
The most pronounced and stage-specific change amongst these variable codons was a marked increase in GAA in the ribosome aminoacyl (A) sites from cells undergoing mitosis. 
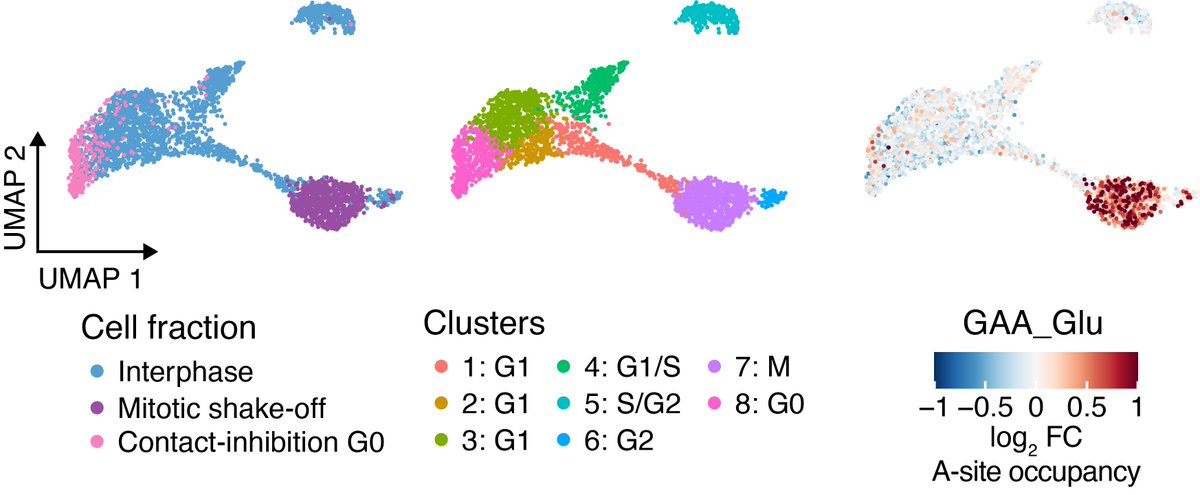
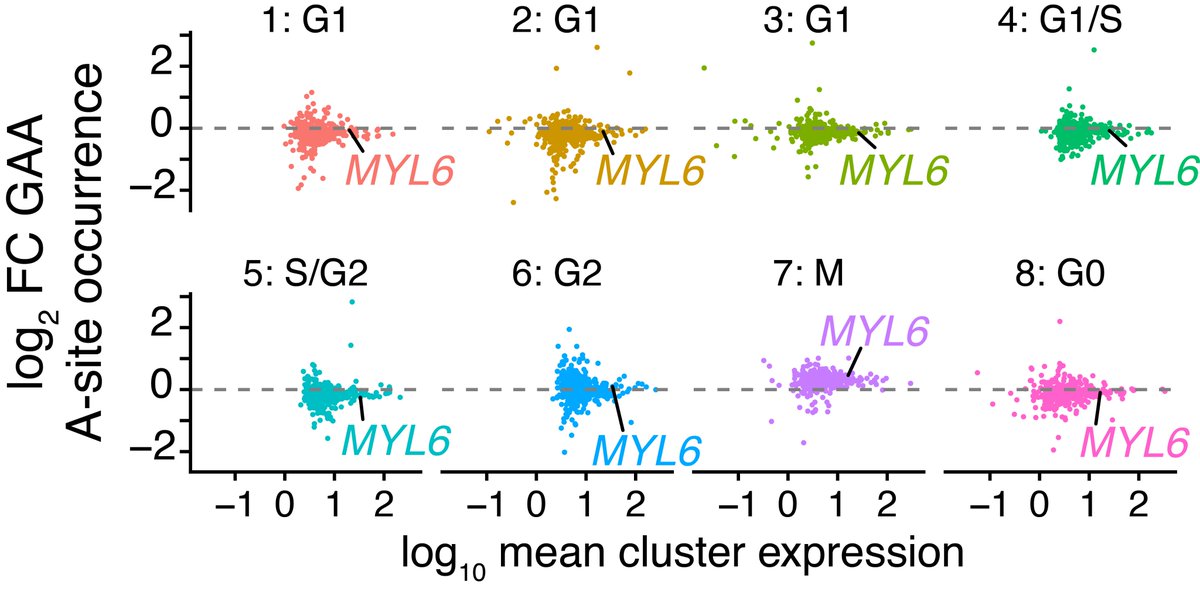
This increase in GAA pausing was further seen to be universal, affecting the majority of translated genes. These stage-specific changes to A-site pausing may reflect global alterations in translation elongation dynamics during mitosis. 
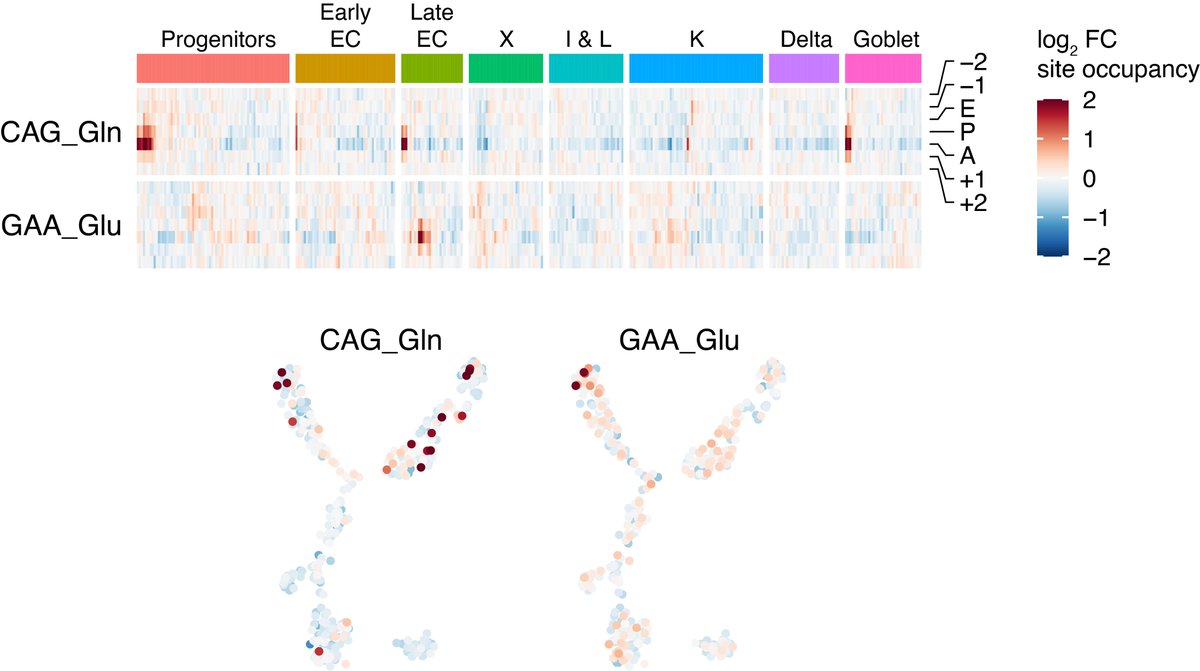
Finally, to demonstrate that scRibo-seq is applicable to complex primary samples, we profiled mouse intestinal enteroendocrine cells. EECs are a rare subpopulation in the gastro-intestinal epithelium that produce and secrete diverse hormones in response to nutrient stimuli
These primary EEC cells displayed the same characteristics of ribosome profiling libraries, and cluster and marker gene analysis revealed that we recovered all the expected EEC cell types in the intestinal crypts. 
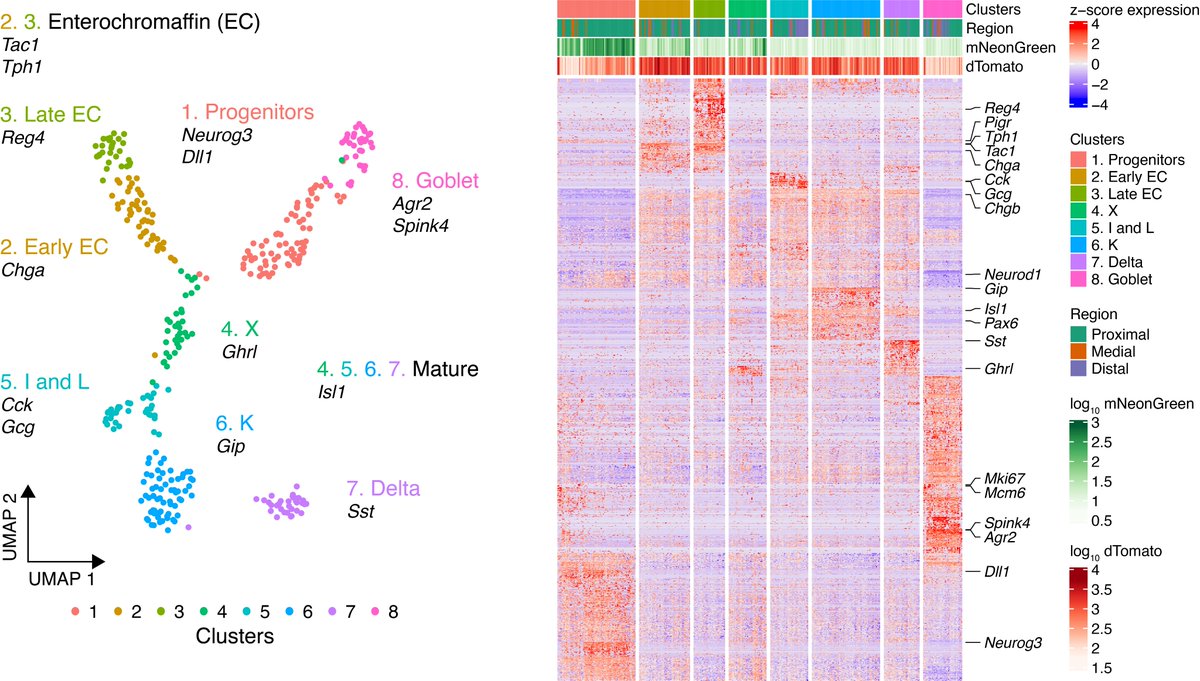
Among the cells were two minority subpopulations that showed genome-wide pausing over CAG and GAA codons, with the GAA-pausing population restricted to the late enterochromaffin cells. 
We are excited to start to apply scRibo-seq to other systems where we hope it will help determine the contribution of the translational process to the astonishing diversity between seemingly identical cells!
• • •
Missing some Tweet in this thread? You can try to
force a refresh



