
New #TrackMate version out (v&7)
With major changes and improvements I describe below (probably a long thread).
It is the product of a great (and cool) collaboration with @guijacquemet lab (@JwPylvanainen), the @DumenilLab lab in @institutpasteur and the IAH facility
1/n
With major changes and improvements I describe below (probably a long thread).
It is the product of a great (and cool) collaboration with @guijacquemet lab (@JwPylvanainen), the @DumenilLab lab in @institutpasteur and the IAH facility
1/n

The new version is the product of an almost Codiv19-induced full rewrite of TrackMate. Our main goal was to allow integrate SOTA segmentation algorithms and tools such as #StarDist, @ilastik_team, #Weka, @MorphoLibJ and #cellpose in TrackMate,
2/n
2/n
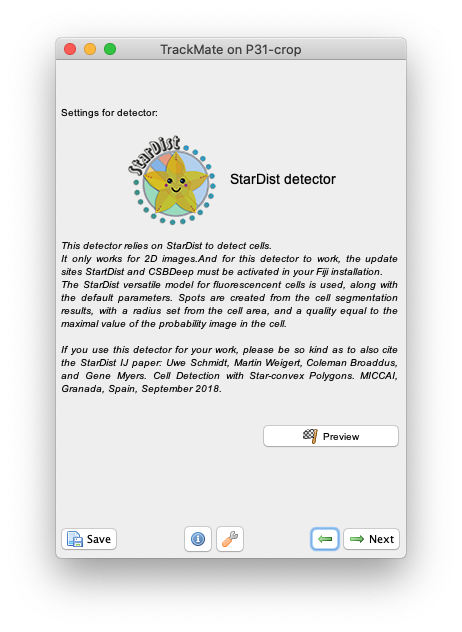
so that they can be used in a tracking pipeline and improve the tracking accuracy and results.
3/n
3/n
So now in TrackMate v7 there are 7 new detectors:
- StarDist, Ilastik, Weka, MorpholibJ on one hand (need to subscripe to a special Fiji update site)
- Detectors for mask images, label images and grayscale images (built-in TrackMate).
4/n
- StarDist, Ilastik, Weka, MorpholibJ on one hand (need to subscripe to a special Fiji update site)
- Detectors for mask images, label images and grayscale images (built-in TrackMate).
4/n
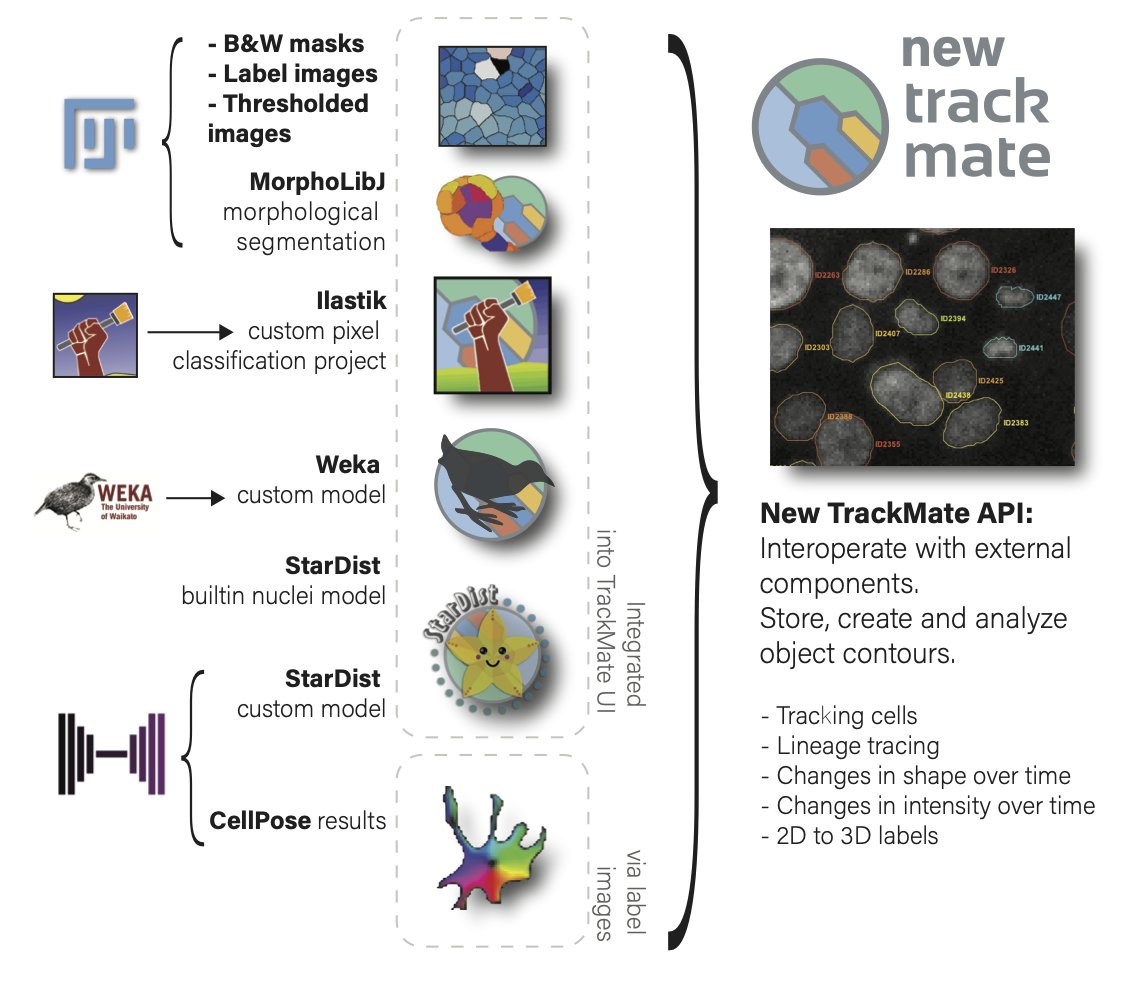
Also! With the new TrackMate version we move from *detection* algorithms (e.g. the LoG detector that has been with TrackMate since its inception) to *segmentation* algorithms, that can return the shape of objects
5/n
5/n

Now TrackMate can exploit this. It supports retrieving, displaying, saving / loading and using the shape of objects. The bad news: only for 2D data.
6/n
6/n
The shapes are used to offer new feature values. For instance there are now morphological features for objects such as area, circularity, ellipse fit etc...
7/n
7/n

And when an object has a shape, TrackMate uses it to compute intensity features (we don't use the spherical shape when we have a contour).
8/n
8/n
There are plenty of other changes we made, touching almost every part of TrackMate. I will maybe speak about that later. But I want to highlight what you can do with the new detectors.
9/n
9/n
For instance #StarDist is awesome when it comes to track dense roundish objects. The fact that we can detect cells accurately thanks to StarDist gives a much more robust tracking results and facilitate the detection of cell division events.
10/n
10/n
The analysis above was made with the built-in StarDist detector in TrackMate, but there is another version that lets you use your own DL model. You can use it to track cells in phase contrast (below, T-cells)
Since we have learned to integrate external algos in TrackMate, we could also integrate #ilastik thanks to the beautiful Fiji tool made by @ilastik_team . It lets you use a pixel classifier in TrackMate.
12/n
12/n
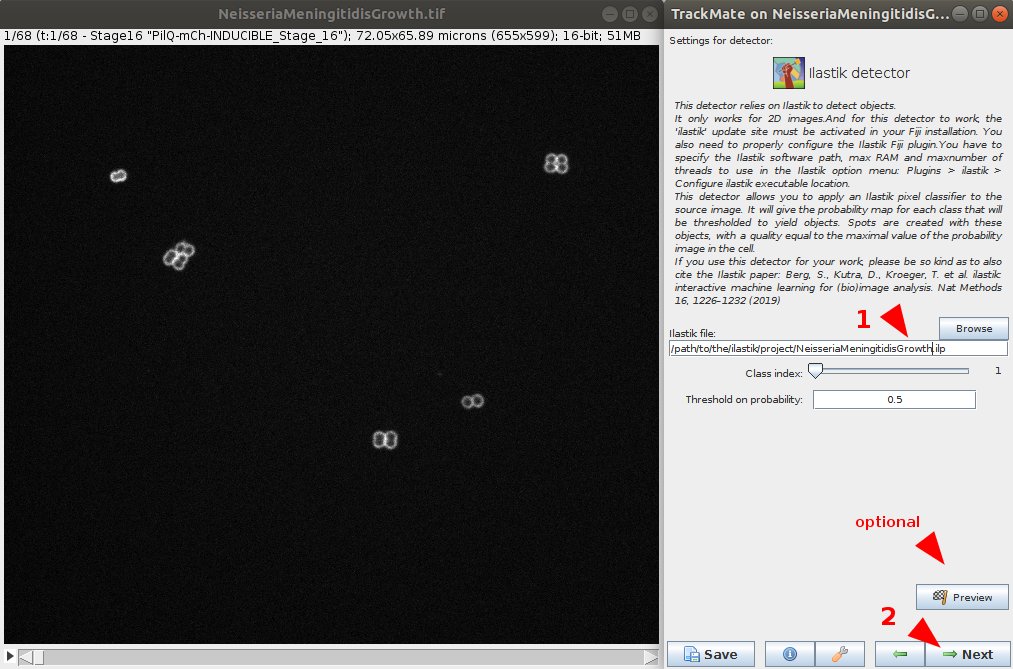
We used it to build the lineages Neisseria meningitidis during bacterial growth (@DumenilLab @StRigaud )
13/n
13/n

So with this kind of analysis, you get for free:
- the lineage of each bacteria with respect with the clone
- the morphological features of each bacteria
- the position of each bacteria with respect to the clone (spatial info)
- and the dynamics feature (speed etc)
14/n
- the lineage of each bacteria with respect with the clone
- the morphological features of each bacteria
- the position of each bacteria with respect to the clone (spatial info)
- and the dynamics feature (speed etc)
14/n
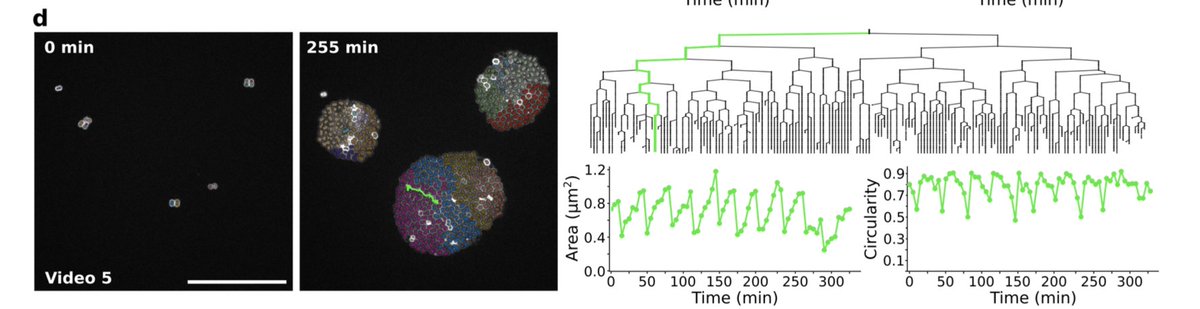
For instance here we follow the area & circularity of a bacteria as it divides. At each cell division we have a sharp decrease in area, that increases quasi linearly in between. The circularity plateaus except just before the division.
Cool no?
15/n
Cool no?
15/n
When it comes to machine learning, Fiji ships the Weka plugin, that we integrated as well. We used for instance to track focal adhesion in cells:
16/n
16/n
And now that we started integrating the beautiful plugin of @IgnacioArganda and friends, we also included his (and David Legland and friends) #MorphoLibJ to track cells stained for their membrane.
(movie by @ksedzinski)
17/n
(movie by @ksedzinski)
17/n
Ok now the big question is what about the rest? What if we are not happy with StarDist, ilastik, MorphoLibJ, Weka? For instance if we want to use #cellpose?
18/n
18/n
Fear not! We did two things:
1. We added general detectors that can use a B&W mask, a label image, or even a grayscale image (e.g. a probability map) as input.
And you can use that to import results from other segmentation algos.
19/n
1. We added general detectors that can use a B&W mask, a label image, or even a grayscale image (e.g. a probability map) as input.
And you can use that to import results from other segmentation algos.
19/n
2/ We made an API so that you can integrate your algorithm YOURSELF!
A key feature of #TrackMate is to be a platform everyone can use to quickly develop their own pipeline.
There is now a new API to let you integrate whatever algo you want in a not too hard way.
21/n
A key feature of #TrackMate is to be a platform everyone can use to quickly develop their own pipeline.
There is now a new API to let you integrate whatever algo you want in a not too hard way.
21/n
For instance, the incredible @haesleinhuepf made a TrackMate detector based on #CLIJ (he managed to do it before TrackMate was released, quite a feat :)
23/n
https://twitter.com/haesleinhuepf/status/1436992074943975429?s=20
23/n
I will stop there but there are so many things that changed! The changelog is here:
github.com/fiji/TrackMate…
24/n
github.com/fiji/TrackMate…
24/n
I loved *every* bit of this work. I got to work with great people. Thank you @ctrueden @jan_eglinger @pietzscht for the support.
25/n
25/n
Have a look at our preprint that describes the new version. There is even a nice analysis of the positive impact you can expect from using DL algos in difficult situation
biorxiv.org/content/10.110…
26/n
biorxiv.org/content/10.110…
26/n
We were careful to write a documentation and tutorials with datasets (on @ZENODO_ORG) to support users. You can browse help from here:
imagej.net/plugins/trackm…
and in the preprint.
27/n
imagej.net/plugins/trackm…
and in the preprint.
27/n
• • •
Missing some Tweet in this thread? You can try to
force a refresh




