
Time for another PK tweetorial, this time featuring a deeper dive on the not-so-mystical art of human dose prediction. 1/ 

I’ve argued previously that human dose is the master optimization parameter in medicinal chemistry. Like death, it can’t be cheated. All variables come together in this one number. 2/ 

This earlier thread talks about some of these things at a higher level and is worth a review. 3/
https://twitter.com/KRHornberger/status/1417442183561547778
Get human dose prediction right: smoother Phase I trial. Get it wrong: your Phase I trial will fail on pharmacokinetics and/or patients will be taking 10 horse pills a day. (nb – that was not an ivermectin joke.) 4/
References first. Repeating two key references that were in the older tweet quoted above. Both of these are gems, and required reading. Also want to re-emphasize, I didn’t invent any of this. I just learned how to use it. 5/
pubs.acs.org/doi/10.1021/mp…
pubs.acs.org/doi/10.1021/mp…
By the end of this, I hope to convince you that medicinal chemists can & should be routinely predicting human dose for lots of compounds during optimization. Basic dose prediction is NOT a specialist activity where you genuflect at the altar and your DMPK person is the priest. 7/
Math alert: as with many other PK topics, it requires some high school algebra to delve into this subject. Also some calculus to derive the equations, which I’ll spare you, as it’s tedious and not relevant to the practical use I want to get to. 8/ 

Let’s dive in. Open your favorite PK text and you’ll find this equation, which relates the concentration of a drug (C) at a time (t) to oral PK parameters: dose (D), bioavailability (F), volume of distribution (Vss), and absorption (ka) and elimination (ke) rate constants. 9/ 

The equation is written this way classically because it’s derived from the solution of a differential equation that looks at concentration as a function of time. But with a bit of algebra, we can rearrange it to solve for the parameter we care about, dose. 10/ 

Now all that’s left, and maybe where chemists get intimidated and decide they want off the ride, is to figure out what all those values are for our compound of interest. “What the f**k is an absorption rate constant? Never seen that for my compound!” is colorful and typical. 11/
Let’s start with concentration and time. This is where it becomes super-important to understand the relationship between PK, PD, and efficacy. How much drug exposure do you need to be efficacious? You must have a critical mass of in vivo data to know that. 12/
Read the thread below if you haven’t already before continuing. It gets into all of this. The dose equation assumes a “time over MEC” scenario, so if you know e.g., that you need to be above the IC50 for 24 hours, there’s your concentration and time. 13/
https://twitter.com/KRHornberger/status/1431235152005447683
But if you read the thread, you know it’s also free drug that matters. The dose equation is rooted in total concentrations (as most PK parameters are measured total rather than unbound), so you’ll also need unbound fraction in human (fu) to convert to total concentration. 14/ 

Next, volume of distribution (Vss). You’ll want Vss data from iv PK in preclinical species. As a first approximation, average these Vss as an estimate for human. Unbound Vss is better to average/scale, but if fu is similar across species, total Vss is an okay approximation. 15/ 

Incidentally, if you’re wondering why Vss matters for oral dose prediction… the answer is, it only matters for time-over-MEC. Vss has a big say in the shape of the PK curve and peak-to-trough ratio. But it matters not in some other scenarios (vide infra). 16/
Next, elimination rate constant (ke). This is scary-sounding at first, but it isn’t. ke is defined as ke = Cl / Vss. Goodbye scary and unfamiliar rate constant, hello familiar iv PK parameters. 17/ 
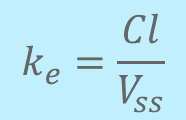
Ah, but now we need to figure out human clearance (Cl). If you’re fortunate, you’ve measured in vitro intrinsic clearance (Cl_int) in e.g. microsomes some preclinical species, and after correcting for fu, discovered that it correlates with an in vivo iv Cl. 18/
If so, great! Your chemical series has an in vitro-in vivo correlation (IVIVC). In such cases, as a first approximation, you can use the intrinsic clearance in e.g. human microsomes to estimate human in vivo clearance (after fu correction). Done and done. 19/ 
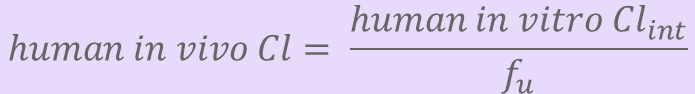
If, however, you’re unfortunate… more experimentation required. You’ll want to measure iv Cl in at least 3 (or better, 4) preclinical species and then go through the gymnastics of allometric scaling. This also sounds scary but isn’t. 20/
All allometry means is there’s a power law relationship between the size of an organism and some physiologic parameter, such as clearance. This means a log-log plot of the physiologic parameter vs. some measure of size will give a straight line. 21/
en.wikipedia.org/wiki/Allometry
en.wikipedia.org/wiki/Allometry
(Fun aside: all sorts of wacky things follow allometric relationships, like the optimal cruising velocity of flying things to their mass to the 1/6 power. Which may finally allow us to put to bed the question of the airspeed velocity of an unladen swallow.) 22/ 

For “simple” allometric scaling of Cl, all that’s required is to plot log Cl vs. log body weight and fit the data to a power law. You can do all of this in Excel, no prob. Then extrapolate the equation up to 70 kg (archetypal human body weight), and there’s your human Cl. 23/ 

A complication: empirically, “simple” allometry is known to overpredict human Cl in many regimes. There are lots of methods out there to unf**k this. A simple one that works well and can be applied with blunt empiricism is the “rule of exponents”. 24/
jpharmsci.org/article/S0022-…
jpharmsci.org/article/S0022-…
All the rule of exponents method says is, depending on the value of the exponent from the curve fit in simple allometry, apply corrections to the allometry to scale by either Cl, Cl * maximum lifespan potential (MLP), or Cl * brain weight. 25/
Yes, that’s some seriously random sh*t to apply corrections with, but it tends to work. Read the paper. For our purposes, suffice to say that MLP and brain weights for species are published, and you can do all those curve fits in Excel too to get a more accurate human Cl. 26/
Moving along… bioavailability (F). First up, it’s best to review this thread on the difference between bioavailability and fraction absorbed before continuing. (All the referral back to old posts shows how ALL this stuff is deeply interlinked.) 27/
https://twitter.com/KRHornberger/status/1426163421616254978
With that reviewed, two ways to proceed. Easy way: you’re making compounds with awesome physchem properties that are generally super well-absorbed. Assume fraction absorbed (technically, fa * fg) = 1 and you can predict human F is the maximum F = 1 – Cl/Qh. 28/ 

Hard way: your series isn’t well-absorbed. Averaging F across preclinical species would be invalid, because it doesn’t correct for species differences in Cl. Instead, average fa (technically, fa * fg) and calculate human F = averaged (fa * fg) * (1 – Cl/Qh). 29/ 

Lastly… that pesky absorption rate constant (ka). As the name implies, this says something about how fast your compound absorbs, and relates to tmax. It comes in units of reciprocal time, just like the elimination rate constant. 30/
Bad news: this one is the most abstract, and tougher to determine from experiment, although there are graphical methods to do so. Good news: it’s one of the most insensitive determinants of dose. A big error here makes a small difference in dose. 31/
For a well-absorbed compound, assuming ka = 1 h-1 is a reasonable starting point. You can empirically slide that down to 0.5, or 0.1, if your tmax in your preclinical species po PK is significantly delayed (say 3-4 h or later). 32/
So let’s recap. That’s a lot of parameters, but nearly all of them are easy to get a handle on. If you have a well-behaved series with an IVIVC and good absorption, all you need to get rolling is: a PK/PD hypothesis, fu, Cl_int from human microsomes, and Vss in 1+ species. 33/
If you have a bad-behaved series with no IVIVC and/or poor absorption, then you need: a PK/PD hypothesis, fu, Cl from 3+ species for allometry, Vss in 1+ species, and fabs in 1+ species. Requires more in vivo PK studies and a more math is the major difference. 34/
Now, before leaving the math behind, I want to leave you with this. If your PK/PD/efficacy relationship is based on AUC and not time over MEC, you’re in luck. That’s a MUCH simpler case. Here’s the equation relating dose and AUC. 35/ 

In this case, all you need is your target AUC, F, and Cl. This is MUCH easier than time over MEC. Why? Because AUC doesn’t depend on things that determine curve shape, so ka, ke, and Vss fall away. If you find yourself here, count your blessings. 36/
Once you have the minimum data set, then it’s just a matter of turning the crank on the dose equation. You can brute-force it with a calculator, write an Excel spreadsheet, or if you’re feeling elegant, a KNIME script. See below (again). 37/
https://twitter.com/KRHornberger/status/1417442197688000513
All of this is well within the capabilities of the garden variety medicinal chemist. Once you grasp the basics, you don’t need a DMPK expert to do this stuff behind the curtain where you don’t even understand where the dose calculations came from. 38/
Now… I sense some DMPK folks out there with twitching eyes. No offense intended; much respect. At the end of the day, you’re still the expert, and none of this thread can or should replace expert guidance when e.g. you’re projecting dose for a regulatory filing. 39/
DMPK experts are awesome. In my experience, they're both exceptionally detail-oriented AND (to chemists) maddeningly imprecise. It’s not a fault – it’s approaching the problem from a radically different perspective than chemists do. 40/
Experts are engaged, and needed, late in the game, when you’re down to your final compound, or couple of compounds. They use sophisticated software to estimate human dose, and they’ll give you a range of possible outcomes – because there’s error in all of the above. 41/
Medicinal chemists, on the other hand… we’re simple creatures. We just want a number. One number. The number from crude human dose predictions may not be 💯% accurate or cover all possible edge cases, but it’s still useful for rank ordering compounds. 42/
Once you get it down, and maybe write some tools to automate the messy bits, it becomes easy to use human dose prediction as an optimization parameter. It liberates you from the ridiculous tyranny of the screening cascade, which can always be gamed. 43/
I’ve also been pleasantly surprised over the years how often the chemist’s crude estimate with a sinfully large bucket of assumptions can get close to expert estimates with very sophisticated modeling tools. Maybe it’s not as mystical as we sometimes feel. /end
Corrigendum: it was pointed out to me by @NoseRubInvest that in post 19, in my haste to put together equation graphics, I inverted the position of Cl and Cl_int. Below is the corrected equation: human in vivo Cl = human in vitrol Cl * fu. Accuracy matters!

https://twitter.com/KRHornberger/status/1438823546172620803

• • •
Missing some Tweet in this thread? You can try to
force a refresh




