
As promised, a 🧵 to convince you to consider SP4 for your next #proteomics sample prep!
TL;DR: Cell lysate -> pure peptides ready for LC-MS with 20-30 min handling time w/o any filters (FASP, iST, S-Trap) or specialist beads (SP3). 1/n
biorxiv.org/content/10.110… @biorxivpreprint
TL;DR: Cell lysate -> pure peptides ready for LC-MS with 20-30 min handling time w/o any filters (FASP, iST, S-Trap) or specialist beads (SP3). 1/n
biorxiv.org/content/10.110… @biorxivpreprint

Starting off a lab (babraham.ac.uk/our-research/s…) where proteomics was going to be a major approach, our first recruit—postdoc @HarveyEJohnston—set off to craft our work-horse sample prep work-flow.
2/n
2/n
This workflow needed to be: (1) quick-ish; (2) cheap-ish; (3) versatile & scalable (e.g., different sample amounts & vols); (4) able to clean up detergents; (5) compatible with #PTM enrichment approaches (especially #ubiquitin TUBEs & diGly); (3) easy to teach to students.
3/n
3/n
After a lot of advice (as founding member of @LPDGproteomics, Harvey has some good connections!) & initial testing, magnetic bead-based SP3 came out as a top contender. nature.com/articles/s4159… embopress.org/doi/full/10.15…
4/n
4/n

Harvey noticed that the use of acetonitrile to promote protein aggregation in SP3 was essentially identical to the protocol (introduced to him by @Rlchardkay) designed to deplete and *exclude* proteins by precipitation for peptidomics – so why not to *capture* proteins?
5/n
5/n
Now, solvent precipitation is a well-worn method for protein isolation & clean-up—developed well before I was born! However, there are 'issues' (e.g., extended incubation, incomplete precipitation/recovery, chemical protein modification from acetone). pubs.acs.org/doi/full/10.10…
6/n
6/n
Recent papers looking SP3 suggest that bead chemistry (i.e., the carboxylate-coating to promote protein capture) is unnecessary—and that protein capture is actually driven by old-school solvent-based precipitation/aggregation.
mcponline.org/article/S1535-…
pubs.acs.org/doi/10.1021/ac…
7/n
mcponline.org/article/S1535-…
pubs.acs.org/doi/10.1021/ac…
7/n

Based on these observations, Harvey went a step further: what if we got rid of the beads entirely, using acetonitrile from the original SP3 protocol to precipitate proteins, and then centrifugation to capture these precipitates?
8/n
8/n
I.e., can the surface chemistry of proteins act in the same way as the surface of the beads?
9/n
9/n
Harvey did a head-to-head comparison of SP3 with this ‘solvent-precipitation SP3’—which we’re calling SP4, to emphasise the similarity and evolution of the method.
10/n
10/n

To understand if bead surface area was also seeding precipitation, Harvey also included a 3rd condition with inert, low-cost (~£0.21/g) glass beads. Spoiler – it was! And the beads helped with digestion too.
11/n
11/n
Evaluating 1 to 5000 μg of protein from a HEK293 lysate, Harvey shows that SP4 (esp. + glass beads) gives equivalent or greater proteome coverage and reproducibility than SP3.
12/n
12/n
The strong similarities between proteomes generated by SP3 and SP4 would suggest (to us anyway!) that protein precipitation is indeed the primary mechanism of SP3.
13/n
13/n
Harvey also found that SP4 is perfectly suited for removing detergents (including SDS, present in the 5-detergent SP3 lysis buffer), and even works with the recently described ‘SPEED’ protocol (lysis & precipitation in 100% TFA).
mcponline.org/article/S1535-…
14/n
mcponline.org/article/S1535-…
14/n
Deep proteome profiling—TMTsixplex, Lys-C+trypsin digestion, followed by off-line peptide fractionation into 62 fractions—showed almost identical results for SP3, SP4 (bead-free), and SP4 (+glass beads), WITH ONE NOTABLE EXCEPTION…
15/n
15/n
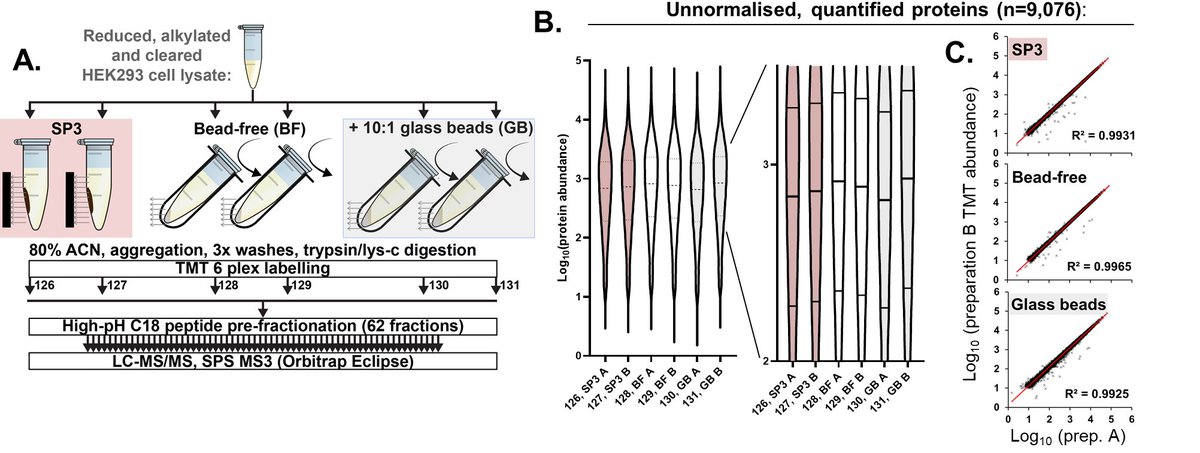
…and that exception was membrane proteins. GO analysis of the small proportion of differentially recovered proteins between SP4 and SP3 revealed an enrichment of membrane proteins.
16/n
16/n

We’re not sure why, but *maybe* (hands waving wildly): highly hydrophobic proteins (such as those with TM domains) aggregate spontaneously in aqueous lysate and thus don’t stick to beads well enough to withstand multiple wash-magnetisation steps.
17/n
17/n
So, proteins that have already aggregated before addition of the magnetic beads will be lost with the washes if they aren’t retained on the bead surface during magnetic capture. (Harvey has a slightly different hypothesis--read the pre-print!)
18/n
18/n
As our group cares a lot about misfolded proteins—which will tend to aggregate in the cell and/or on lysis—we think SP4 (or any centrifugation-based capture) will be superior to magnetic beads for our work. We’re currently testing this in various ways (stay tuned!).
19/n
19/n
And while we’re talking about misfolded proteins, we’ve also tried this with diGly ubiquitin remnant profiling—and it works! We need to validate this application a bit more, and are saving it for a juicy story coming soon (hopefully…).
20/n
20/n
Anyway, back to what we DO show here: To make sure that he wasn’t missing something, Harvey shared the step-by-step protocol (included as a Supplement in the pre-print) with a few colleagues (@BabrahamInst's MS core, Holger Kramer @MRC_LMS, and Jo Kirkpatrick @TheCrick).
21/n
21/n
To our delight-relief, all our co-authors came back smiling, i.e., similar if not slightly better performance with SP4 vs. SP3. Thanks to Kranthi, David, Jo, @georgebiggs94, & Holger for your enthusiastic support and work!
22/n
22/n

Anyway, that’s it! Happy to field any comments or Qs. Or simply give it a go! SP4 came from very rigorous R&D by Harvey for proteostasis-ageing biology projects kicking off in the lab. That’s what we’re off to do now, but hopefully SP4 is useful for other labs too. Cheers!
23/n
23/n
• • •
Missing some Tweet in this thread? You can try to
force a refresh



