Since the @UN @UNBiodiversity was discussing biodiversity targets this week, we wanted to post our latest study in @biorxiv_ecology
“Quantifying the scale of genetic diversity extinction in the Anthropocene”
doi.org/10.1101/2021.1…
@CarnegiePlants @CarnegieEcology @Stanford
🧵
“Quantifying the scale of genetic diversity extinction in the Anthropocene”
doi.org/10.1101/2021.1…
@CarnegiePlants @CarnegieEcology @Stanford
🧵
This all started with a deep worry about our lack of knowledge of global losses of genetic diversity and the call for more conservation policies that account for genetics @LindaLaikre, @seanmhoban, and others 

in summer 2021, a new @UN press release (motivated by a piece by @sdiazecology 10.1126/science.abe1530) presented the goal to preserve “at least 90% of genetic diversity within all species”. Great news, but how do we get there and measure progress?
un.org/sustainabledev…
un.org/sustainabledev…

The direct approach to track progress should work: we SEQUENCE ALL THE GENOMES AND TRACK GENETIC DIVERSITY over time, but empirical studies show shy signals (Leight et al Evo App, Millette et al Eco Letter). Perhaps there's not enough (genomic and temporal) data yet!
in the meantime we started thinking about how genetic diversity is lost as species geographic distributions shrink due to global change. We dug into math biodiversity theory used for biodiversity risk assessments and translated it into population genetic theory
(Cute fact: R.A. Fisher’s log-Series to model species abundances is a general version of the Site Frequency Spectrum 1/q in popgen, also by Fisher (under some conditions). To keep theoreticians also interested 🧐)
We also downloaded genomic datasets till we passed the mark of 10K individuals and a total of almost 20 plant and animal species (don’t forget about plants! 🌱 dx.doi.org/10.1111/tpj.14…) with continental geo-tagged smpling so we could test our theory
We describe the Mutations-Area Relationship (MAR), something that feels intuitively obvious, but this has not explicitly parameterized (that we know): essentially, if you randomly reduce the geographic area of a species (done by simulations), what is the loss of gen. diversity? 

(MAR is a weird popgen acronym; we could use genetics area relationship or something similar but MAR in Spanish means “sea” and that is nice to think about every time we write about it, reminds me of my hometown Alicante by the Mediterranean) 

Finally, multiple back-of-the-envelope calculations using our theory and global ecosystem transformations based on remote sensing point to over 10% genetic diversity losses worldwide, a substantial amount!! 

We realize there is a lot of uncertainty, so instead of focusing on a single estimate we mapped the possible values of genetic extinction with this surface plot where we have all possible ecosystems area transformations and MAR uncertainty. 
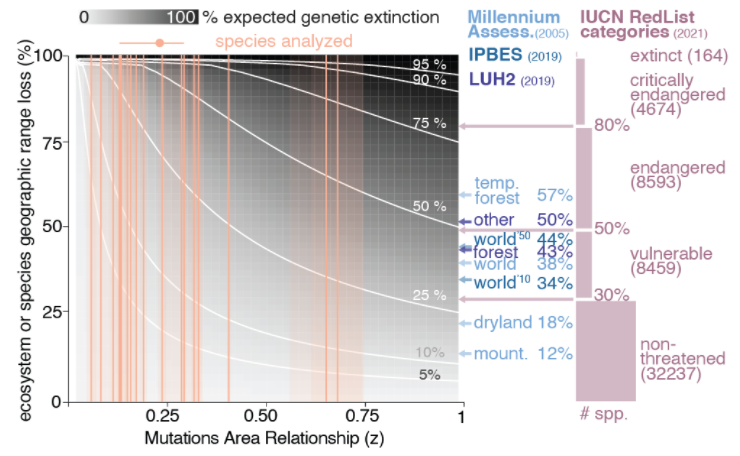
(note: the word "extinction" applied to genetics may sound odd, and we could use "loss". this is an intended metaphor to emphasize that, because the rate of genetic diversity recovery is much slower than the losses, this loss is effectively irreversible. check the supp materials)
given that our work predicts we are close to (or already) failing @UNBiodiversity target of 90% genetic diversity protection across all species, we need to aggressively plan for protection of populations and germplasms from many geographic enclaves! our method can help plan! 

This preprint is perhaps the most important work we have done to contribute to understanding global change impacts on biodiversity, and is also a working draft that we’d love to get feedback on! **We need to get this right, so please feel free to DM me!**
🙏thankful to an amazing team and collaborators
@TomBooker12 @LucCzech @TadashiFukami @leg2015 @s_hateley @ChrisKyriazis @plantricia @laura_leventhal @Noguesbravo @VPagowski @MeganRRuffley @spence_jeffrey_ seba @clw_gg @ZessingAround
@TomBooker12 @LucCzech @TadashiFukami @leg2015 @s_hateley @ChrisKyriazis @plantricia @laura_leventhal @Noguesbravo @VPagowski @MeganRRuffley @spence_jeffrey_ seba @clw_gg @ZessingAround
• • •
Missing some Tweet in this thread? You can try to
force a refresh