Having this enormous collection of pQTLs allows us to answer the question (again):
Which is more relevant:
Distance of a GWAS SNP to the TSS (transcription start site) or to the gene body of a candidate gene?
pubmed.ncbi.nlm.nih.gov/34648354/
Which is more relevant:
Distance of a GWAS SNP to the TSS (transcription start site) or to the gene body of a candidate gene?
pubmed.ncbi.nlm.nih.gov/34648354/
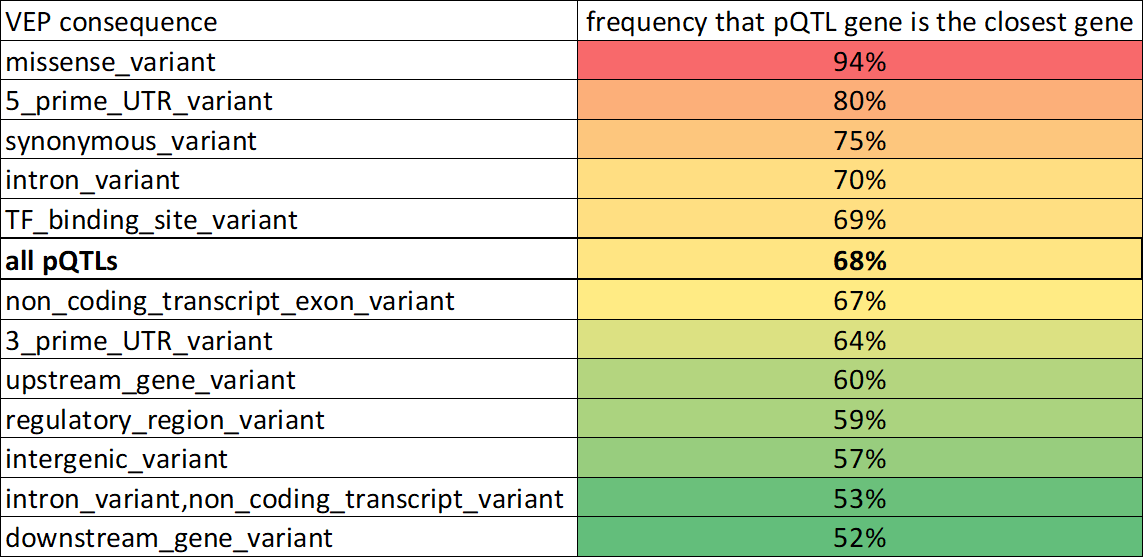
Usually you get the same closest gene measuring to TSS or to gene body.
But in the top case a pQTL for ACAA1 sits inside an irrelevant gene but is closer to the TSS for ACAA1.
But a pQTL for DNAJC17 sits closer to a TSS for a random gene despite sitting within DNAJC17.
But in the top case a pQTL for ACAA1 sits inside an irrelevant gene but is closer to the TSS for ACAA1.
But a pQTL for DNAJC17 sits closer to a TSS for a random gene despite sitting within DNAJC17.

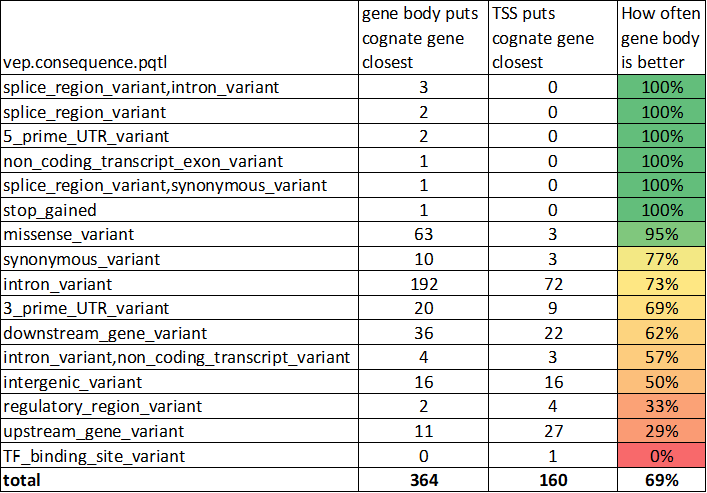
Turns out if TSS_closest_gene and gene_body_closest_gene disagree, the gene body metric is right twice as often
This is especially true if the SNP sits within a gene, even if it is not a missense variant
(Again though, usually TSS and gene_body agree on the closest gene (77%))
This is especially true if the SNP sits within a gene, even if it is not a missense variant
(Again though, usually TSS and gene_body agree on the closest gene (77%))
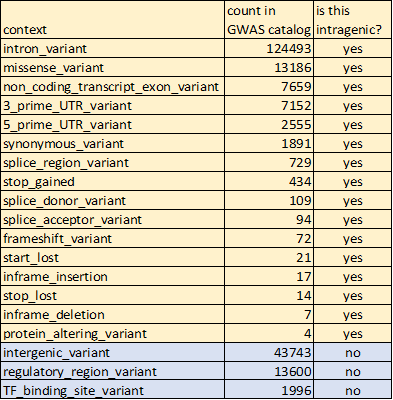
This is important because MOST GWAS SNPs SIT WITHIN A GENE. That is, even though most GWAS SNPs are non-coding, they are not intergenic.
This table counts how often each "context" term is used in the @GWASCatalog
where p<5e-8 and a context is provided.
This table counts how often each "context" term is used in the @GWASCatalog
where p<5e-8 and a context is provided.
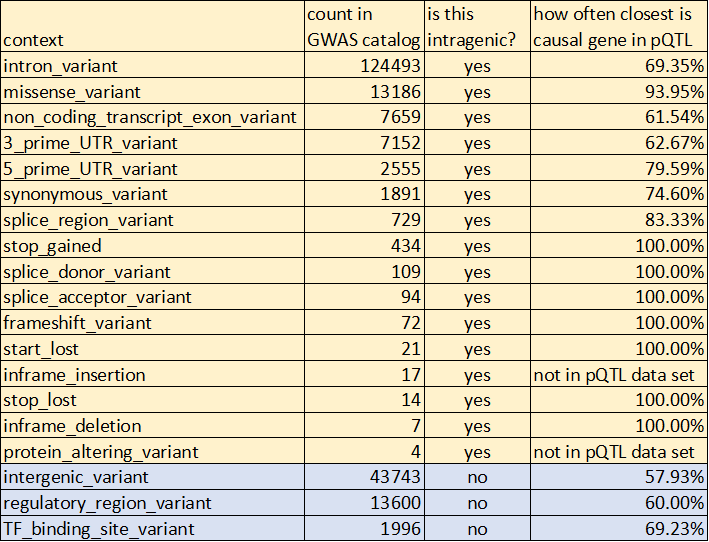
Combining @GWASCatalog tally with pQTL data from @pietznerm et al:
* Most GWAS SNPs (all traits) sit within a gene; that gene is ~70% likely to be the causal gene (pQTL data)
* Intergenic SNPs are only 27% of GWAS SNPs; here closest gene is causal 59%
* expected weighted avg =68%
* Most GWAS SNPs (all traits) sit within a gene; that gene is ~70% likely to be the causal gene (pQTL data)
* Intergenic SNPs are only 27% of GWAS SNPs; here closest gene is causal 59%
* expected weighted avg =68%
• • •
Missing some Tweet in this thread? You can try to
force a refresh