
I am happy to conclude my trilogy on Cas9 specificity with our new preprint from the @MartinJinek lab! We solved a staggering number (15!) of crystal structures of Cas9 bound to bona fide off-targets to investigate the nature of mismatch tolerance.
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…

We observe that mismatch tolerance is primarily facilitated by the formation of non-canonical base pairs within the heteroduplex. This effect is dependent on the type of mismatch, the surrounding nucleotides, and its position within the duplex. 

As the level of protein coordination varies along the duplex, in some cases we see that preservation of proper base stacking is preferred over unfavourable pairing. We also observe HNH side-chain coordination of an rC-dT mismatch (on-target positioning in white)! 

The PAM-proximal region of the target strand allows for backbone flexibility which can result in some rather exotic base pairs, such as an rA-dG in Watson-Crick orientation. This, however, can misposition the scissile phosphate, providing another level of off-target regulation. 
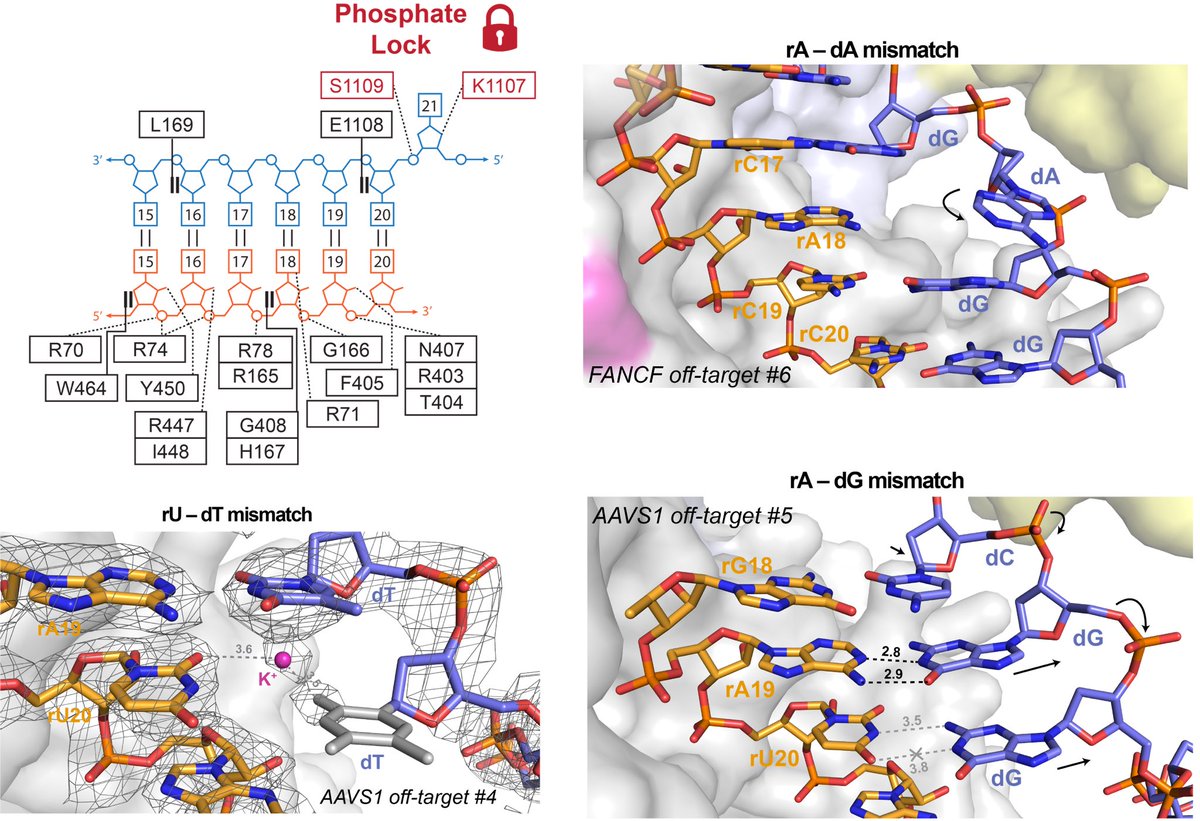
We also observe two cases of RNA base skipping to accommodate off-targets containing predicted single nucleotide deletions. This is preferred to the previously proposed “base bulging out” mechanism as it allows to maintain a decent degree of base stacking. 
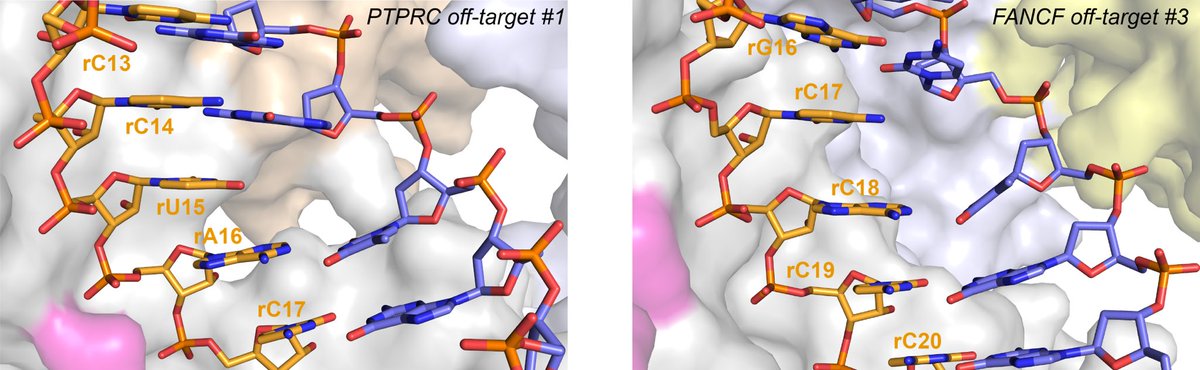
We propose that this can only occur within the seed region where the RNA strand is heavily coordinated by Cas9. In an off-target with a predicted single nucleotide deletion in the PAM-distal part we instead observe tolerance of consecutive multiple mismatches (on-target white). 
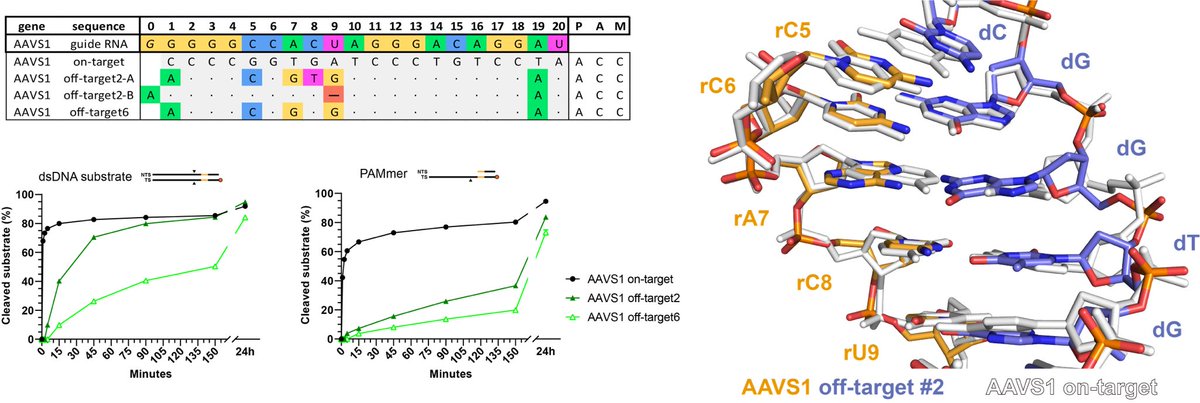
Strikingly, by reversing one of the mismatches and thereby reducing the total amount of mismatches, we actually observe severely reduced cleavage of this offtarget! This implies that mismatches can have synergistic effects on heteroduplex conformation and Cas9 activity!
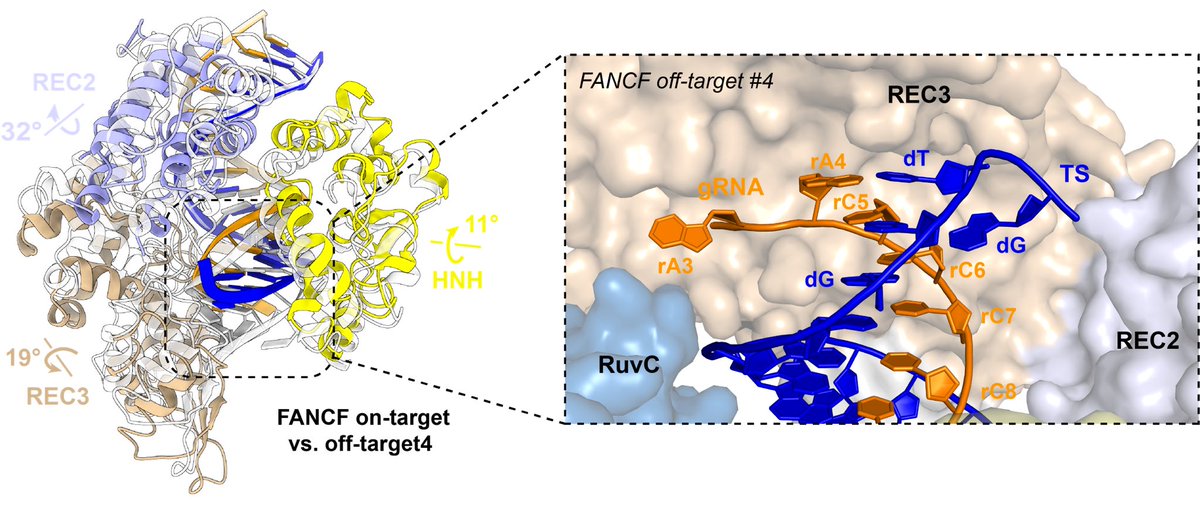
Lastly we solve the structure of an off-target containing mismatches in the last three PAM-distal positions, mimicking a truncated RNA. This heteroduplex unpairing induces a REC-lobe rearrangement, similar to the one observed in xCas9 3.7 or RNA-DNA hybrid guided complexes. 
I’d like to thank @CaribouBio, specifically Paul Donohoue and Peter Cameron for their support with off-target profiling. Also I would like to thank @AcleryClery and Frederic Allain for assaying binding kinetics. Lastly, big thanks to Katja Bargsten for preparing LOADS of buffers
I am very happy to discuss our results and conclusions. The large amount and diversity of structures has made analysis challenging, so I am very interested to hear new thoughts and ideas!
Here is a morph movie between the on-target bound and off-target bound structure with PAM-distal mismatches to highlight the rearrangement of the REC2/3 and HNH domains relative to the heteroduplex.
We also observe some flexibility of the REC2 domain, when we compare the AAVS1 on-target bound structure and the FANCF/TRAC on-target bound structures.
• • •
Missing some Tweet in this thread? You can try to
force a refresh



