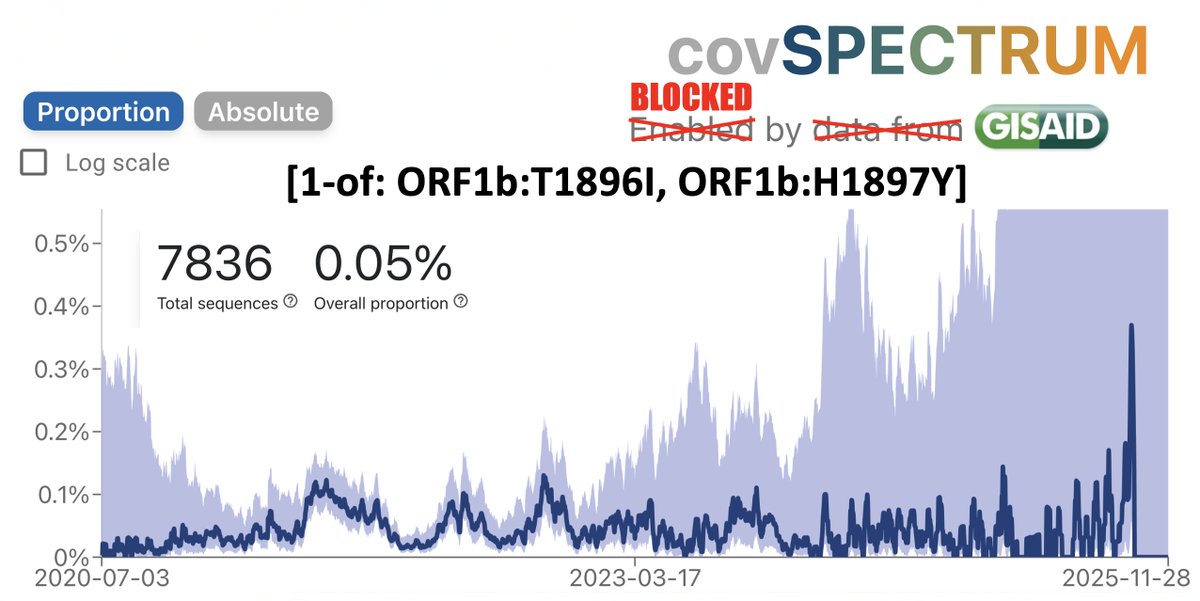
1/6 Imagine that: Omicron is undergoing phenomenally fast exponential growth in the US, just like in the UK, Denmark, and South Africa. Crossing the Atlantic didn't impair its transmissibility. Who could've guessed?
Dec 6—3%
Dec 7—7%
Dec 8—13%
Dec 6—3%
Dec 7—7%
Dec 8—13%
https://twitter.com/ByMikeBaker/status/1470536653252882434

2/6 Take it from the master himself, Trevor B: "There is an inevitable very large wave of Omicron. It's going to happen." nytimes.com/live/2021/12/1… 

3/6 There's such an air of fatalism around all this, as if we are utterly incapable of doing anything that could dampen or avert a devastating Omicron wave. Hospitals are already at max capacity in many states. An Omicron tsunami approaches, & we collectively shrug our shoulders.
4/6 Epidemiologist @sanghyuk_shin of UC Irvine: "We need to take this seriously, starting now. If we have learned anything on how this virus operates—it’s that any kind of mitigation, the earlier the better...." voiceofoc.org/2021/12/local-… 

5/6 "...There is really no evidence that suggests that Omicron is going to be mild, there’s no evidence that it is less virulent." 

6/6 At a company Christmas party at an Oslo restaurant, 80 out of 111 young (ages 30-50), 2-dose vaccinated Norwegians were infected with Omicron. Only 1 of the 80 was asymptomatic (none hospitalized). I've never heard of an asymptomatic rate so low. It doesn't suggest mildness. 

• • •
Missing some Tweet in this thread? You can try to
force a refresh